[English] 日本語
 Yorodumi
Yorodumi- SASDFJ8: Glutamate/aspartate import solute-binding protein (DEBP) in the p... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Glutamate/aspartate import solute-binding protein (DEBP) in the presence of glutamate - Glu-bound 10-fold excess
|
| Function / homology |  Function and homology information Function and homology informationaspartate binding / L-glutamate transmembrane transport / L-aspartate transmembrane transport / L-aspartate import across plasma membrane / L-glutamate import across plasma membrane / glutamate binding / amino acid transport / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / outer membrane-bounded periplasmic space / extracellular region / membrane Similarity search - Function |
| Biological species |  |
 Citation Citation |  Date: 2019 Aug 1 Date: 2019 Aug 1Title: Structure-based screening of binding affinities via small-angle X-ray scattering Authors: Chen P / Masiewicz P / Perez K |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: Glutamate/aspartate import solute-binding protein (DEBP) in the presence of glutamate - Glu-bound 10-fold excess Specimen concentration: 2.2 mg/ml |
|---|---|
| Buffer | Name: 100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP / pH: 7.4 / Comment: SEC buffer for PBP experiments |
| Entity #1755 | Type: protein Description: Glutamate/aspartate import solute-binding protein Formula weight: 32.052 / Num. of mol.: 1 / Source: Escherichia coli (strain K12) / References: UniProt: P37902 Sequence: HHHHHHDDAA PAAGSTLDKI AKNGVIVVGH RESSVPFSYY DNQQKVVGYS QDYSNAIVEA VKKKLNKPDL QVKLIPITSQ NRIPLLQNGT FDFECGSTTN NVERQKQAAF SDTIFVVGTR LLTKKGGDIK DFANLKDKAV VVTSGTTSEV LLNKLNEEQK MNMRIISAKD ...Sequence: HHHHHHDDAA PAAGSTLDKI AKNGVIVVGH RESSVPFSYY DNQQKVVGYS QDYSNAIVEA VKKKLNKPDL QVKLIPITSQ NRIPLLQNGT FDFECGSTTN NVERQKQAAF SDTIFVVGTR LLTKKGGDIK DFANLKDKAV VVTSGTTSEV LLNKLNEEQK MNMRIISAKD HGDSFRTLES GRAVAFMMDD ALLAGERAKA KKPDNWEIVG KPQSQEAYGC MLRKDDPQFK KLMDDTIAQV QTSGEAEKWF DKWFKNPIPP KNLNMNFELS DEMKALFKEP NDKALN |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.09919 Å / Dist. spec. to detc.: 2.849 mm / Type of source: X-ray synchrotron / Wavelength: 0.09919 Å / Dist. spec. to detc.: 2.849 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | ||||||||||||||||||||||||||||||
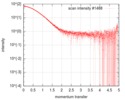
| Scan | Measurement date: Sep 10, 2018 / Cell temperature: 20 °C / Exposure time: 2 sec. / Number of frames: 12 / Unit: 1/nm /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result | Comments: Glu-bound DEBP at a ligand:protein ratio of 10:1. Ligand condition is prepared by dissolving glutamate in powder form (Sigma-Aldrich) directly into the SAXS buffer, followed by dilution to ...Comments: Glu-bound DEBP at a ligand:protein ratio of 10:1. Ligand condition is prepared by dissolving glutamate in powder form (Sigma-Aldrich) directly into the SAXS buffer, followed by dilution to the target concentration with additional SAXS buffer. Note that the scattering intensity therefore contains Na+ counterions that was not directly subtracted.
|
 Movie
Movie Controller
Controller


 SASDFJ8
SASDFJ8