+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Histidine-binding periplasmic protein (HisBP), apo-form
|
| Function / homology |  Function and homology information Function and homology informationL-histidine import across plasma membrane / amino acid binding / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / outer membrane-bounded periplasmic space / membrane Similarity search - Function |
| Biological species |  |
 Citation Citation |  Date: 2019 Aug 1 Date: 2019 Aug 1Title: Structure-based screening of binding affinities via small-angle X-ray scattering Authors: Chen P / Masiewicz P / Perez K |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: Histidine-binding periplasmic protein (HisBP), apo-form Specimen concentration: 4.25 mg/ml |
|---|---|
| Buffer | Name: 100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP / pH: 7.4 / Comment: SEC buffer for PBP experiments |
| Entity #1753 | Name: HisBP / Type: protein / Description: Histidine-binding periplasmic protein / Formula weight: 26.289 / Num. of mol.: 1 / Source: Escherichia coli (strain K12) / References: UniProt: P0AEU0 Sequence: GAIPQNIRIG TDPTYAPFES KNSQGELVGF DIDLAKELCK RINTQCTFVE NPLDALIPSL KAKKIDAIMS SLSITEKRQQ EIAFTDKLYA ADSRLVVAKN SDIQPTVESL KGKRVGVLQG TTQETFGNEH WAPKGIEIVS YQGQDNIYSD LTAGRIDAAF QDEVAASEGF ...Sequence: GAIPQNIRIG TDPTYAPFES KNSQGELVGF DIDLAKELCK RINTQCTFVE NPLDALIPSL KAKKIDAIMS SLSITEKRQQ EIAFTDKLYA ADSRLVVAKN SDIQPTVESL KGKRVGVLQG TTQETFGNEH WAPKGIEIVS YQGQDNIYSD LTAGRIDAAF QDEVAASEGF LKQPVGKDYK FGGPSVKDEK LFGVGTGMGL RKEDNELREA LNKAFAEMRA DGTYEKLAKK YFDFDVYGG |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.09919 Å / Dist. spec. to detc.: 2.849 mm / Type of source: X-ray synchrotron / Wavelength: 0.09919 Å / Dist. spec. to detc.: 2.849 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | |||||||||||||||||||||||||||||||||
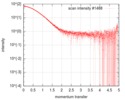
| Scan | Measurement date: Sep 10, 2018 / Cell temperature: 20 °C / Exposure time: 2 sec. / Number of frames: 12 / Unit: 1/nm /
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result | Comments: Free HisBP after TEV cleavage to remove hexahistidine tags, followed by reverse His-Trap and SEC column purification. Note N-terminal glycine before the Uniprot sequence due to TEV tag.
|
 Movie
Movie Controller
Controller



 SASDFD8
SASDFD8