[English] 日本語
 Yorodumi
Yorodumi- SASDD66: Phox homologue (PX) - C2 domains of human phosphatidylinositol 4-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDD66 |
|---|---|
 Sample Sample | Phox homologue (PX) - C2 domains of human phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha (PI3KC2α)
|
| Function / homology |  Function and homology information Function and homology informationvascular associated smooth muscle contraction / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the early endosome membrane / phosphatidylinositol-4-phosphate 3-kinase / Synthesis of PIPs at the Golgi membrane / clathrin coat assembly / phosphatidylinositol biosynthetic process / membrane organization / 1-phosphatidylinositol-4-phosphate 3-kinase activity / phosphatidylinositol-3-phosphate biosynthetic process ...vascular associated smooth muscle contraction / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the early endosome membrane / phosphatidylinositol-4-phosphate 3-kinase / Synthesis of PIPs at the Golgi membrane / clathrin coat assembly / phosphatidylinositol biosynthetic process / membrane organization / 1-phosphatidylinositol-4-phosphate 3-kinase activity / phosphatidylinositol-3-phosphate biosynthetic process / clathrin-coated vesicle / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / positive regulation of cell migration involved in sprouting angiogenesis / phosphatidylinositol-4,5-bisphosphate 3-kinase / phosphatidylinositol 3-kinase / 1-phosphatidylinositol-3-kinase activity / clathrin binding / phosphatidylinositol-mediated signaling / Golgi Associated Vesicle Biogenesis / platelet-derived growth factor receptor signaling pathway / Synthesis of PIPs at the plasma membrane / exocytosis / phosphatidylinositol binding / positive regulation of autophagy / phosphatidylinositol 3-kinase/protein kinase B signal transduction / trans-Golgi network / epidermal growth factor receptor signaling pathway / endocytosis / insulin receptor signaling pathway / cell migration / Clathrin-mediated endocytosis / vesicle / intracellular membrane-bounded organelle / extracellular exosome / nucleoplasm / ATP binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Structure / Year: 2018 Journal: Structure / Year: 2018Title: Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2α. Authors: Kai-En Chen / Vikas A Tillu / Mintu Chandra / Brett M Collins /  Abstract: Phosphorylation of phosphoinositides by the class II phosphatidylinositol 3-kinase (PI3K) PI3K-C2α is essential for many processes, including neuroexocytosis and formation of clathrin-coated ...Phosphorylation of phosphoinositides by the class II phosphatidylinositol 3-kinase (PI3K) PI3K-C2α is essential for many processes, including neuroexocytosis and formation of clathrin-coated vesicles. A defining feature of the class II PI3Ks is a C-terminal module composed of phox-homology (PX) and C2 membrane interacting domains; however, the mechanisms that control their specific cellular localization remain poorly understood. Here we report the crystal structure of the C2 domain of PI3K-C2α in complex with the phosphoinositide head-group mimic inositol hexaphosphate, revealing two distinct pockets for membrane binding. The C2 domain preferentially binds to phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol (3,4,5)-trisphosphate, and low-resolution structures of the combined PX-C2 module by small-angle X-ray scattering reveal a compact conformation in which cooperative lipid binding by each domain binding can occur. Finally, we demonstrate an unexpected role for calcium in perturbing the membrane interactions of the PX-C2 module, which we speculate may be important for regulating the activity of PI3K-C2α. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDD66 SASDD66 |
|---|
-Related structure data
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
| Model #1921 |  Type: dummy / Software: (5.0) / Radius of dummy atoms: 2.25 A / Chi-square value: 0.162  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #1957 |  Type: dummy / Radius of dummy atoms: 2.40 A / Chi-square value: 0.162  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #1922 |  Type: mix / Radius of dummy atoms: 2.00 A / Chi-square value: 0.16  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Phox homologue (PX) - C2 domains of human phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha (PI3KC2α) Specimen concentration: 10.5 mg/ml |
|---|---|
| Buffer | Name: 25 mM Tris 200 mM NaCl 5% Glycerol 0.5 mM TCEP / pH: 8.5 |
| Entity #1041 | Name: PI3KC2α / Type: protein Description: Phox Homology (PX) - C2 domains of human Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha Formula weight: 32.663 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: O00443 Sequence: SNADEPILSF SPKTYSFRQD GRIKEVSVFT YHKKYNPDKH YIYVVRILRE GQIEPSFVFR TFDEFQELHN KLSIIFPLWK LPGFPNRMVL GRTHAKDVAA KRKIELNSYL QSLMNASTDV AECDLVCTFF HPLLRDEKAE GIARSADAGS FSPTPGQIGG AVKLSISYRN ...Sequence: SNADEPILSF SPKTYSFRQD GRIKEVSVFT YHKKYNPDKH YIYVVRILRE GQIEPSFVFR TFDEFQELHN KLSIIFPLWK LPGFPNRMVL GRTHAKDVAA KRKIELNSYL QSLMNASTDV AECDLVCTFF HPLLRDEKAE GIARSADAGS FSPTPGQIGG AVKLSISYRN GTLFIMVMHI KDLVTEDGAD PNPYVKTYLL PDNHKTSKRK TKISRKTRNP TFNEMLVYSG YSKETLRQRE LQLSVLSAES LRENFFLGGV TLPLKDFNLS KETVKWYQLT AATYL |
-Experimental information
| Beam | Instrument name: Australian Synchrotron SAXS/WAXS / City: Melbourne / 国: Australia  / Shape: Point / Type of source: X-ray synchrotron / Wavelength: 0.10332 Å / Shape: Point / Type of source: X-ray synchrotron / Wavelength: 0.10332 Å | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | |||||||||||||||||||||||||||||||||
| Scan | Measurement date: Oct 20, 2017 / Cell temperature: 10 °C / Exposure time: 1 sec. / Number of frames: 14 / Unit: 1/A /
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
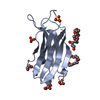
| Result | Comments: SEC-SAXS details: Column type: S200 5/150 GL column; Flow rate: 0.45 ml/min; Sample temperature, 10°C; Sample injection concentration: 10.5 mg/ml. The ab initio models represent the ...Comments: SEC-SAXS details: Column type: S200 5/150 GL column; Flow rate: 0.45 ml/min; Sample temperature, 10°C; Sample injection concentration: 10.5 mg/ml. The ab initio models represent the spatially aligned and volume occupancy corrected (averaged) representation of the protein obtained from 20 individual reconstructions (top, DAMFILT model: NSD = 0.66 +/- 0.02; resolution estimate = 3.3 nm) and the best-fit individual DAMMIN reconstruction (bottom) displayed with the corresponding individual model fit to the SAXS data.
|
 Movie
Movie Controller
Controller