+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Proline utilization A from Desulfovibrio vulgaris 1.5 mg/mL
|
| Function / homology |  Function and homology information Function and homology informationproline dehydrogenase / proline dehydrogenase activity / oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor / L-glutamate gamma-semialdehyde dehydrogenase / L-glutamate gamma-semialdehyde dehydrogenase activity / : / : / DNA-binding transcription factor activity / DNA binding Similarity search - Function |
| Biological species |  Desulfovibrio vulgaris (strain RCH1) (bacteria) Desulfovibrio vulgaris (strain RCH1) (bacteria) |
 Citation Citation |  Journal: FEBS J / Year: 2017 Journal: FEBS J / Year: 2017Title: Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure. Authors: David A Korasick / Harkewal Singh / Travis A Pemberton / Min Luo / Richa Dhatwalia / John J Tanner /  Abstract: Many enzymes form homooligomers, yet the functional significance of self-association is seldom obvious. Herein, we examine the connection between oligomerization and catalytic function for proline ...Many enzymes form homooligomers, yet the functional significance of self-association is seldom obvious. Herein, we examine the connection between oligomerization and catalytic function for proline utilization A (PutA) enzymes. PutAs are bifunctional enzymes that catalyze both reactions of proline catabolism. Type A PutAs are the smallest members of the family, possessing a minimal domain architecture consisting of N-terminal proline dehydrogenase and C-terminal l-glutamate-γ-semialdehyde dehydrogenase modules. Type A PutAs form domain-swapped dimers, and in one case (Bradyrhizobium japonicum PutA), two of the dimers assemble into a ring-shaped tetramer. Whereas the dimer has a clear role in substrate channeling, the functional significance of the tetramer is unknown. To address this question, we performed structural studies of four-type A PutAs from two clades of the PutA tree. The crystal structure of Bdellovibrio bacteriovorus PutA covalently inactivated by N-propargylglycine revealed a fold and substrate-channeling tunnel similar to other PutAs. Small-angle X-ray scattering (SAXS) and analytical ultracentrifugation indicated that Bdellovibrio PutA is dimeric in solution, in contrast to the prediction from crystal packing of a stable tetrameric assembly. SAXS studies of two other type A PutAs from separate clades also suggested that the dimer predominates in solution. To assess whether the tetramer of B. japonicum PutA is necessary for catalytic function, a hot spot disruption mutant that cleanly produces dimeric protein was generated. The dimeric variant exhibited kinetic parameters similar to the wild-type enzyme. These results implicate the domain-swapped dimer as the core structural and functional unit of type A PutAs. ENZYMES: Proline dehydrogenase (EC 1.5.5.2); l-glutamate-γ-semialdehyde dehydrogenase (EC 1.2.1.88). DATABASES: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession number 5UR2. The SAXS data have been deposited in the SASBDB under the ...DATABASES: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession number 5UR2. The SAXS data have been deposited in the SASBDB under the following accession codes: SASDCP3 (BbPutA), SASDCQ3 (DvPutA 1.5 mg·mL ), SASDCX3 (DvPutA 3.0 mg·mL ), SASDCY3 (DvPutA 4.5 mg·mL ), SASDCR3 (LpPutA 3.0 mg·mL ), SASDCV3 (LpPutA 5.0 mg·mL ), SASDCW3 (LpPutA 8.0 mg·mL ), SASDCS3 (BjPutA 2.3 mg·mL ), SASDCT3 (BjPutA 4.7 mg·mL ), SASDCU3 (BjPutA 7.0 mg·mL ), SASDCZ3 (R51E 2.3 mg·mL ), SASDC24 (R51E 4.7 mg·mL ), SASDC34 (R51E 7.0 mg·mL ). |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDCQ3 SASDCQ3 |
|---|
-Related structure data
| Related structure data |  5ur2C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Proline utilization A from Desulfovibrio vulgaris 1.5 mg/mL Specimen concentration: 1.5 mg/ml |
|---|---|
| Buffer | Name: 50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, and 0.5 mM THP at pH 7.5. pH: 7.5 |
| Entity #679 | Name: DvPutA / Type: protein / Description: Bifunctional protein PutA / Formula weight: 114.496 / Num. of mol.: 2 / Source: Desulfovibrio vulgaris (strain RCH1) / References: UniProt: A0A0E0T6R2 Sequence: MHHHHHHSSG VDLGTENLYF QSMDQQHLDG KVVERGKEFF RSISGEAPSI FNKGWWTGKV MDWAMQNEDF KVQLFRFVDV LPYLNTSESL LRHIREYFAT EDADIPPVLK WGAGKAGIGG ALTAKLMGMT IRSNIEGMAR QFIIGDNSKE AVKGLAKLRK DGFTFTVDLL ...Sequence: MHHHHHHSSG VDLGTENLYF QSMDQQHLDG KVVERGKEFF RSISGEAPSI FNKGWWTGKV MDWAMQNEDF KVQLFRFVDV LPYLNTSESL LRHIREYFAT EDADIPPVLK WGAGKAGIGG ALTAKLMGMT IRSNIEGMAR QFIIGDNSKE AVKGLAKLRK DGFTFTVDLL GEATVSEEES EAYAQGYHEV VDAIAREQEK WKALPGNGPV EGFDWGATPK VNVSIKPSAL YSQAKPVDVE GSVRGILSRL VPIYRKVVAM GGFLCIDMEQ LKYKEMTLEL FKRLRSDPEF RHYPHLSIVL QAYLRDTEKD LDDLLHWARS EKLPIGIRLV KGAYWDYETV IAKQNGWEIP VWTDKPESDI AYEKLAHRIL ENSDIVYFAC ASHNVRTIAA VMETALALNV PEHRYEFQVL YGMAEPVRKG LKNVAGRVRL YCPYGELIPG MAYLVRRLLE NTANESFLRQ SFAEGAALER LLENPQKTLH RLLAARPEPR AVEPGPGGLP PFTNDAMIDF TVPDNRKAFV EALADVRSRF GQTVPLYIGG RDVTTADLIP TTNPAKPAEV VASICQAGRP EIDDAIAAAK KAALTWRDTS PADRAAYLRR AADICRKRIW ELSAWQVVEV GKQWDQAYHD VTEGIDFLEY YAREMLRLGA PRRMGRAPGE HNHLFYQPKG IAAVIAPWNF PFAIAIGMAS AAIVTGNPVI FKPSSISSRI GYNLAEVFRE AGLPEGVFNY CPGRSSIMGD YLVEHPDISL ICFTGSMEVG LRIQEKAAKV QPGQRQCKRV IAEMGGKNAT IIDDDADLDE AVLQVLYSAF GFQGQKCSAC SRVIVLDAIY DRFIERLVKA ASSIHIGPSE DPSNYMGPVA DATLQKNVSD YIRIAEEEGR VLLKRTDLPA EGCYVPLTIV GDIRPEHRIA QEEIFGPVLA VMRAATFDEA LSIANGTRFA LTGAVFSRSP EHLDKARREF RVGNLYLNKG STGALVERQP FGGFAMSGVG SKTGGPDYLL QFMDPRVVTE NTMRRGFTPI DEDDDWIV |
-Experimental information
| Beam | Instrument name: Advanced Light Source (ALS) 12.3.1 (SIBYLS) City: Berkeley, CA / 国: USA  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: MAR 165 CCD | |||||||||||||||||||||||||||||||||
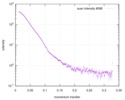
| Scan | Measurement date: Jun 8, 2012 / Unit: 1/A /
| |||||||||||||||||||||||||||||||||
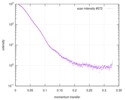
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller