[English] 日本語
 Yorodumi
Yorodumi- SASDC24: Proline utilization A from Bradyrhizobium diazoefficiens (formerl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Proline utilization A from Bradyrhizobium diazoefficiens (formerly Bradyrhizobium japonicum) R51E mutant 4.7 mg/mL
|
| Function / homology |  Function and homology information Function and homology informationproline dehydrogenase / proline dehydrogenase activity / L-glutamate gamma-semialdehyde dehydrogenase / L-glutamate gamma-semialdehyde dehydrogenase activity / : / : / cytoplasmic side of plasma membrane / DNA-binding transcription factor activity / nucleotide binding / DNA binding ...proline dehydrogenase / proline dehydrogenase activity / L-glutamate gamma-semialdehyde dehydrogenase / L-glutamate gamma-semialdehyde dehydrogenase activity / : / : / cytoplasmic side of plasma membrane / DNA-binding transcription factor activity / nucleotide binding / DNA binding / identical protein binding / cytoplasm Similarity search - Function |
| Biological species |  Bradyrhizobium diazoefficiens (strain JCM 10833 / IAM 13628 / NBRC 14792 / USDA 110) (bacteria) Bradyrhizobium diazoefficiens (strain JCM 10833 / IAM 13628 / NBRC 14792 / USDA 110) (bacteria) |
 Citation Citation |  Journal: FEBS J / Year: 2017 Journal: FEBS J / Year: 2017Title: Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure. Authors: David A Korasick / Harkewal Singh / Travis A Pemberton / Min Luo / Richa Dhatwalia / John J Tanner /  Abstract: Many enzymes form homooligomers, yet the functional significance of self-association is seldom obvious. Herein, we examine the connection between oligomerization and catalytic function for proline ...Many enzymes form homooligomers, yet the functional significance of self-association is seldom obvious. Herein, we examine the connection between oligomerization and catalytic function for proline utilization A (PutA) enzymes. PutAs are bifunctional enzymes that catalyze both reactions of proline catabolism. Type A PutAs are the smallest members of the family, possessing a minimal domain architecture consisting of N-terminal proline dehydrogenase and C-terminal l-glutamate-γ-semialdehyde dehydrogenase modules. Type A PutAs form domain-swapped dimers, and in one case (Bradyrhizobium japonicum PutA), two of the dimers assemble into a ring-shaped tetramer. Whereas the dimer has a clear role in substrate channeling, the functional significance of the tetramer is unknown. To address this question, we performed structural studies of four-type A PutAs from two clades of the PutA tree. The crystal structure of Bdellovibrio bacteriovorus PutA covalently inactivated by N-propargylglycine revealed a fold and substrate-channeling tunnel similar to other PutAs. Small-angle X-ray scattering (SAXS) and analytical ultracentrifugation indicated that Bdellovibrio PutA is dimeric in solution, in contrast to the prediction from crystal packing of a stable tetrameric assembly. SAXS studies of two other type A PutAs from separate clades also suggested that the dimer predominates in solution. To assess whether the tetramer of B. japonicum PutA is necessary for catalytic function, a hot spot disruption mutant that cleanly produces dimeric protein was generated. The dimeric variant exhibited kinetic parameters similar to the wild-type enzyme. These results implicate the domain-swapped dimer as the core structural and functional unit of type A PutAs. ENZYMES: Proline dehydrogenase (EC 1.5.5.2); l-glutamate-γ-semialdehyde dehydrogenase (EC 1.2.1.88). DATABASES: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession number 5UR2. The SAXS data have been deposited in the SASBDB under the ...DATABASES: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession number 5UR2. The SAXS data have been deposited in the SASBDB under the following accession codes: SASDCP3 (BbPutA), SASDCQ3 (DvPutA 1.5 mg·mL ), SASDCX3 (DvPutA 3.0 mg·mL ), SASDCY3 (DvPutA 4.5 mg·mL ), SASDCR3 (LpPutA 3.0 mg·mL ), SASDCV3 (LpPutA 5.0 mg·mL ), SASDCW3 (LpPutA 8.0 mg·mL ), SASDCS3 (BjPutA 2.3 mg·mL ), SASDCT3 (BjPutA 4.7 mg·mL ), SASDCU3 (BjPutA 7.0 mg·mL ), SASDCZ3 (R51E 2.3 mg·mL ), SASDC24 (R51E 4.7 mg·mL ), SASDC34 (R51E 7.0 mg·mL ). |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDC24 SASDC24 |
|---|
-Related structure data
| Related structure data |  5ur2C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Proline utilization A from Bradyrhizobium diazoefficiens (formerly Bradyrhizobium japonicum) R51E mutant 4.7 mg/mL Specimen concentration: 4.7 mg/ml |
|---|---|
| Buffer | Name: 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine, and 5% (v/v) glycerol pH: 7.8 / Comment: BjPutA buffer |
| Entity #682 | Name: BjPutA R51E / Type: protein / Description: Proline dehydrogenase / Formula weight: 107.531 / Num. of mol.: 2 Source: Bradyrhizobium diazoefficiens (strain JCM 10833 / IAM 13628 / NBRC 14792 / USDA 110) References: UniProt: Q89E26 Sequence: GHMPNIPPPF TAPYAPDDAE IAARLLPASH LSPPQEARIH RTATRLIEAI RKEDDRLGGV EDMLREFALS TKEGLALMVL AEALLRVPDA RTADQFIEDK LGEGDFIHHE TKSTAFLVNA SAWALGLSAR VIQPGETPDG TIGRLVKRLG APAVRTATRQ AMRLMGNHFV ...Sequence: GHMPNIPPPF TAPYAPDDAE IAARLLPASH LSPPQEARIH RTATRLIEAI RKEDDRLGGV EDMLREFALS TKEGLALMVL AEALLRVPDA RTADQFIEDK LGEGDFIHHE TKSTAFLVNA SAWALGLSAR VIQPGETPDG TIGRLVKRLG APAVRTATRQ AMRLMGNHFV LGETIEQALE RGKPRSGQKT RYSFDMLGEG ARTAADARRY FDAYASAIET IGKAAGNHAL PDRPGISVKL SALHPRFEAI SRARVMVELV PQLLDLAQRA KAHDLNFTVD AEEADRLELS LDVIAATLAD PSLKGWDGFG LAIQAYQKRA SAVIDYVDAL ARAHDRKLMV RLVKGAYWDT EIKRAQERGL DGYPVFTRKA MTDLNYVACA SKLLALRPRI FPQFATHNAL TVATVLEMAE GSSGFEFQRL HGMGEALYEQ LAKDHADIAY RTYAPVGSHR DLLAYLVRRL LENGANSSFV AQAADYRVPV PALLQRPADA IVRPQAAAHP RIPLPCDLFA PERRNSRGVE FGARTALDQL LTDVKAETGD LKPIADATPD QAHAAVAAAR AGFAGWSRTP AGIRAAALEQ AAHLLESRSA HFIALLQREG GKTLDDALSE LREAADFCRY YAAQGRKLFG SETAMPGPTG ESNALTMRGR GVFVAISPWN FPLAIFLGQV TAALMAGNSV VAKPAEQTPR IAREAVALLH EAGIPKSALY LVTGDGRIGA ALTAHPDIAG VVFTGSTEVA RSINRALAAK DGPIVPLIAE TGGINAMIAD ATALPEQVAD DVVTSAFRSA GQRCSALRLL FVQEDVADRM IEMVAGAARE LKIGDPSDVA THVGPVIDVE AKQRLDAHIA RMKTEARLHF AGPAPEGCFV APHIFELTEA GQLTEEVFGP ILHVVRYRPE NLERVLRAIE RTGYGLTLGV HSRIDDSIEA IIDRVQVGNI YVNRNMIGAV VGVQPFGGNG LSGTGPKAGG PHYLARFATE QTVTINTAAA GGNAALLAGE E |
-Experimental information
| Beam | Instrument name: Advanced Light Source (ALS) 12.3.1 (SIBYLS) City: Berkeley, CA / 国: USA  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus3 X 2M / Pixsize x: 172 mm | |||||||||||||||||||||||||||||||||
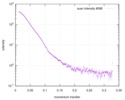
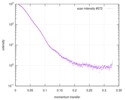
| Scan | Measurement date: Dec 16, 2016 / Unit: 1/A /
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller