[English] 日本語
 Yorodumi
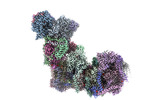
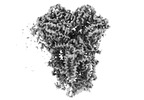
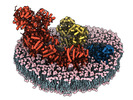
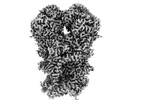
Yorodumi- PDB-8ugp: In-situ structure of typeA supercomplex in respiratory chain ( lo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8ugp | ||||||
|---|---|---|---|---|---|---|---|
| Title | In-situ structure of typeA supercomplex in respiratory chain ( local refined map focused on CI iron-sulfur cluster regions ) | ||||||
 Components Components |
| ||||||
 Keywords Keywords | ELECTRON TRANSPORT / in-situ cryo-EM structure / mammalian / mitochondria / respiratory supercomplex / proton pumping / membrane protein | ||||||
| Function / homology |  Function and homology information Function and homology informationRHOG GTPase cycle / Complex I biogenesis / Respiratory electron transport / Mitochondrial protein degradation / cellular respiration / ubiquinone-6 biosynthetic process / oxidoreductase activity, acting on NAD(P)H / NADH:ubiquinone reductase (H+-translocating) / apoptotic mitochondrial changes / : ...RHOG GTPase cycle / Complex I biogenesis / Respiratory electron transport / Mitochondrial protein degradation / cellular respiration / ubiquinone-6 biosynthetic process / oxidoreductase activity, acting on NAD(P)H / NADH:ubiquinone reductase (H+-translocating) / apoptotic mitochondrial changes / : / mitochondrial electron transport, NADH to ubiquinone / mitochondrial respiratory chain complex I assembly / NADH dehydrogenase (ubiquinone) activity / quinone binding / electron transport coupled proton transport / negative regulation of intrinsic apoptotic signaling pathway / ATP metabolic process / response to cAMP / aerobic respiration / reactive oxygen species metabolic process / regulation of mitochondrial membrane potential / electron transport chain / regulation of protein phosphorylation / mitochondrial intermembrane space / brain development / negative regulation of cell growth / NAD binding / positive regulation of fibroblast proliferation / 4 iron, 4 sulfur cluster binding / response to oxidative stress / mitochondrial inner membrane / nuclear body / protein-containing complex binding / mitochondrion / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2 Å | ||||||
 Authors Authors | Zheng, W. / Zhang, K. / Zhu, J. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: High-resolution in situ structures of mammalian respiratory supercomplexes. Authors: Wan Zheng / Pengxin Chai / Jiapeng Zhu / Kai Zhang /   Abstract: Mitochondria play a pivotal part in ATP energy production through oxidative phosphorylation, which occurs within the inner membrane through a series of respiratory complexes. Despite extensive in ...Mitochondria play a pivotal part in ATP energy production through oxidative phosphorylation, which occurs within the inner membrane through a series of respiratory complexes. Despite extensive in vitro structural studies, determining the atomic details of their molecular mechanisms in physiological states remains a major challenge, primarily because of loss of the native environment during purification. Here we directly image porcine mitochondria using an in situ cryo-electron microscopy approach. This enables us to determine the structures of various high-order assemblies of respiratory supercomplexes in their native states. We identify four main supercomplex organizations: IIIIIV, IIIIIV, IIIIIV and IIIIIV, which potentially expand into higher-order arrays on the inner membranes. These diverse supercomplexes are largely formed by 'protein-lipids-protein' interactions, which in turn have a substantial impact on the local geometry of the surrounding membranes. Our in situ structures also capture numerous reactive intermediates within these respiratory supercomplexes, shedding light on the dynamic processes of the ubiquinone/ubiquinol exchange mechanism in complex I and the Q-cycle in complex III. Structural comparison of supercomplexes from mitochondria treated under different conditions indicates a possible correlation between conformational states of complexes I and III, probably in response to environmental changes. By preserving the native membrane environment, our approach enables structural studies of mitochondrial respiratory supercomplexes in reaction at high resolution across multiple scales, from atomic-level details to the broader subcellular context. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8ugp.cif.gz 8ugp.cif.gz | 207.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8ugp.ent.gz pdb8ugp.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8ugp.json.gz 8ugp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8ugp_validation.pdf.gz 8ugp_validation.pdf.gz | 1.5 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8ugp_full_validation.pdf.gz 8ugp_full_validation.pdf.gz | 1.5 MB | Display | |
| Data in XML |  8ugp_validation.xml.gz 8ugp_validation.xml.gz | 40.8 KB | Display | |
| Data in CIF |  8ugp_validation.cif.gz 8ugp_validation.cif.gz | 56.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ug/8ugp https://data.pdbj.org/pub/pdb/validation_reports/ug/8ugp ftp://data.pdbj.org/pub/pdb/validation_reports/ug/8ugp ftp://data.pdbj.org/pub/pdb/validation_reports/ug/8ugp | HTTPS FTP |
-Related structure data
| Related structure data |  42231MC  8ud1C  8ueoC  8uepC  8ueqC  8uerC  8uesC  8uetC  8ueuC  8uevC  8uewC  8uexC  8ueyC  8uezC  8ugdC  8ugeC  8ugfC  8uggC  8ughC  8ugiC  8ugjC  8ugkC  8uglC  8ugnC  8ugrC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-NADH-ubiquinone oxidoreductase chain ... , 2 types, 2 molecules 1A1H
| #1: Protein | Mass: 12998.448 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #6: Protein | Mass: 35591.426 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  References: UniProt: A0A0U1RS44, NADH:ubiquinone reductase (H+-translocating) |
-NADH dehydrogenase [ubiquinone] iron-sulfur protein ... , 6 types, 6 molecules 1B1C1D1I1Q1R
| #2: Protein | Mass: 28500.184 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #3: Protein | Mass: 30154.318 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #4: Protein | Mass: 54168.074 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #7: Protein | Mass: 27135.039 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #9: Protein | Mass: 19746.514 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #10: Protein | Mass: 13348.950 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein , 1 types, 1 molecules 1G
| #5: Protein | Mass: 79627.398 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit ... , 3 types, 3 molecules 1P1W1q
| #8: Protein | Mass: 42606.324 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #11: Protein | Mass: 14953.313 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #12: Protein | Mass: 17162.439 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Non-polymers , 2 types, 4 molecules 


| #13: Chemical | | #14: Chemical | ChemComp-ZN / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: In-situ structure of typeA supercomplex in respiratory chain ( local refined map focused on CI iron-sulfur cluster regions ) Type: COMPLEX / Entity ID: #1-#12 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:  |
| Buffer solution | pH: 7 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 3000 nm / Nominal defocus min: 1300 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Version: 1.20.1_4487: / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 60000 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj











