+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7onb | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of the U2 5' module of the A3'-SSA complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SPLICING / Spliceosome / spliceostatin A / splicing inhibitor | ||||||
| Function / homology |  Function and homology information Function and homology informationU11/U12 snRNP / B-WICH complex / U12-type spliceosomal complex / RNA splicing, via transesterification reactions / splicing factor binding / U2-type precatalytic spliceosome / U2-type prespliceosome assembly / U2-type spliceosomal complex / SAGA complex / U2 snRNP ...U11/U12 snRNP / B-WICH complex / U12-type spliceosomal complex / RNA splicing, via transesterification reactions / splicing factor binding / U2-type precatalytic spliceosome / U2-type prespliceosome assembly / U2-type spliceosomal complex / SAGA complex / U2 snRNP / U2-type prespliceosome / positive regulation of transcription by RNA polymerase III / precatalytic spliceosome / regulation of RNA splicing / spliceosomal complex assembly / mRNA 3'-splice site recognition / positive regulation of transcription by RNA polymerase I / mRNA Splicing - Minor Pathway / U2 snRNA binding / regulation of DNA repair / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / RNA splicing / stem cell differentiation / spliceosomal complex / positive regulation of neuron projection development / mRNA splicing, via spliceosome / negative regulation of protein catabolic process / B-WICH complex positively regulates rRNA expression / nuclear matrix / mRNA processing / nuclear speck / chromatin remodeling / mRNA binding / positive regulation of DNA-templated transcription / protein-containing complex binding / nucleolus / positive regulation of transcription by RNA polymerase II / DNA binding / RNA binding / zinc ion binding / nucleoplasm / nucleus Similarity search - Function | ||||||
| Biological species |  unidentified adenovirus unidentified adenovirus Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||
 Authors Authors | Cretu, C. / Pena, V. | ||||||
| Funding support |  Germany, 1items Germany, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors. Authors: Constantin Cretu / Patricia Gee / Xiang Liu / Anant Agrawal / Tuong-Vi Nguyen / Arun K Ghosh / Andrew Cook / Melissa Jurica / Nicholas A Larsen / Vladimir Pena /    Abstract: Intron selection during the formation of prespliceosomes is a critical event in pre-mRNA splicing. Chemical modulation of intron selection has emerged as a route for cancer therapy. Splicing ...Intron selection during the formation of prespliceosomes is a critical event in pre-mRNA splicing. Chemical modulation of intron selection has emerged as a route for cancer therapy. Splicing modulators alter the splicing patterns in cells by binding to the U2 snRNP (small nuclear ribonucleoprotein)-a complex chaperoning the selection of branch and 3' splice sites. Here we report crystal structures of the SF3B module of the U2 snRNP in complex with spliceostatin and sudemycin FR901464 analogs, and the cryo-electron microscopy structure of a cross-exon prespliceosome-like complex arrested with spliceostatin A. The structures reveal how modulators inactivate the branch site in a sequence-dependent manner and stall an E-to-A prespliceosome intermediate by covalent coupling to a nucleophilic zinc finger belonging to the SF3B subunit PHF5A. These findings support a mechanism of intron recognition by the U2 snRNP as a toehold-mediated strand invasion and advance an unanticipated drug targeting concept. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7onb.cif.gz 7onb.cif.gz | 540.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7onb.ent.gz pdb7onb.ent.gz | 382.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7onb.json.gz 7onb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/on/7onb https://data.pdbj.org/pub/pdb/validation_reports/on/7onb ftp://data.pdbj.org/pub/pdb/validation_reports/on/7onb ftp://data.pdbj.org/pub/pdb/validation_reports/on/7onb | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  12994MC  7b0iC  7b91C  7b92C  7b9cC  7omfC  7opiC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Splicing factor 3B subunit ... , 5 types, 5 molecules ABCIK
| #1: Protein | Mass: 135718.844 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q15393 Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q15393 |
|---|---|
| #2: Protein | Mass: 10149.369 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q9BWJ5 Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q9BWJ5 |
| #3: Protein | Mass: 146024.938 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: O75533 Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: O75533 |
| #8: Protein | Mass: 100377.812 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q13435 Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q13435 |
| #11: Protein | Mass: 44436.570 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q15427 Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q15427 |
-RNA chain , 2 types, 2 molecules GH
| #6: RNA chain | Mass: 73393.234 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  unidentified adenovirus / Production host: synthetic construct (others) unidentified adenovirus / Production host: synthetic construct (others) |
|---|---|
| #7: RNA chain | Mass: 60186.445 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: GenBank: 36516 Homo sapiens (human) / Cell line: HeLa S3 / References: GenBank: 36516 |
-Splicing factor 3A subunit ... , 2 types, 2 molecules MN
| #9: Protein | Mass: 49327.355 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q15428 Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q15428 |
|---|---|
| #10: Protein | Mass: 58934.844 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q12874 Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q12874 |
-Protein / Protein/peptide , 2 types, 2 molecules DF
| #4: Protein | Mass: 12427.524 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q7RTV0 Homo sapiens (human) / Cell line: HeLa S3 / References: UniProt: Q7RTV0 |
|---|---|
| #5: Protein/peptide | Mass: 1999.216 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / Cell line: HeLa S3 Homo sapiens (human) / Cell line: HeLa S3 |
-Non-polymers , 2 types, 6 molecules 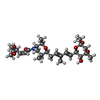


| #12: Chemical | ChemComp-SJT / |
|---|---|
| #13: Chemical | ChemComp-ZN / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | ||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||
| Source (recombinant) | Organism: synthetic construct (others) | ||||||||||||||||||||||||
| Buffer solution | pH: 7.9 | ||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||
| Specimen | Conc.: 0.24 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid type: UltrAuFoil R1.2/1.3 | ||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K Details: The A3'-SSA complex was frozen in vitreous ice by applying ~2.2 uL crosslinked sample to both sides of a glow-discharged UltrAufoil R1.2/1.3 gold grid. The excess sample was blotted away for ...Details: The A3'-SSA complex was frozen in vitreous ice by applying ~2.2 uL crosslinked sample to both sides of a glow-discharged UltrAufoil R1.2/1.3 gold grid. The excess sample was blotted away for 2s using a blot force of 7 and the grid was snap frozen in liquid ethane cooled by liquid nitrogen. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 81000 X / Calibrated magnification: 81000 X / Nominal defocus max: 2500 nm / Nominal defocus min: 1000 nm / Cs: 2.7 mm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 2 sec. / Electron dose: 41.41 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 10494 |
| EM imaging optics | Energyfilter name: GIF Quantum LS / Energyfilter slit width: 20 eV |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1052001 | ||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 78262 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: OTHER / Space: REAL | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building |
| ||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 127.46 Å2 | ||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj





































