[English] 日本語
 Yorodumi
Yorodumi- PDB-7b92: Structure of a minimal SF3B core in complex with sudemycin D6 (fo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7b92 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a minimal SF3B core in complex with sudemycin D6 (form II) | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | SPLICING / SF3B / pre-mRNA splicing / splicing modulator / sudemycin D6 | |||||||||
| Function / homology |  Function and homology information Function and homology informationU11/U12 snRNP / B-WICH complex / U12-type spliceosomal complex / RNA splicing, via transesterification reactions / splicing factor binding / U2-type precatalytic spliceosome / U2-type prespliceosome assembly / U2-type spliceosomal complex / SAGA complex / U2 snRNP ...U11/U12 snRNP / B-WICH complex / U12-type spliceosomal complex / RNA splicing, via transesterification reactions / splicing factor binding / U2-type precatalytic spliceosome / U2-type prespliceosome assembly / U2-type spliceosomal complex / SAGA complex / U2 snRNP / U2-type prespliceosome / positive regulation of transcription by RNA polymerase III / precatalytic spliceosome / regulation of RNA splicing / spliceosomal complex assembly / positive regulation of transcription by RNA polymerase I / mRNA Splicing - Minor Pathway / U2 snRNA binding / regulation of DNA repair / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / RNA splicing / stem cell differentiation / spliceosomal complex / mRNA splicing, via spliceosome / negative regulation of protein catabolic process / B-WICH complex positively regulates rRNA expression / nuclear matrix / nuclear speck / chromatin remodeling / mRNA binding / positive regulation of DNA-templated transcription / protein-containing complex binding / nucleolus / positive regulation of transcription by RNA polymerase II / DNA binding / RNA binding / zinc ion binding / nucleoplasm / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3 Å MOLECULAR REPLACEMENT / Resolution: 3 Å | |||||||||
 Authors Authors | Cretu, C. / Pena, V. | |||||||||
| Funding support |  Germany, 1items Germany, 1items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors. Authors: Constantin Cretu / Patricia Gee / Xiang Liu / Anant Agrawal / Tuong-Vi Nguyen / Arun K Ghosh / Andrew Cook / Melissa Jurica / Nicholas A Larsen / Vladimir Pena /    Abstract: Intron selection during the formation of prespliceosomes is a critical event in pre-mRNA splicing. Chemical modulation of intron selection has emerged as a route for cancer therapy. Splicing ...Intron selection during the formation of prespliceosomes is a critical event in pre-mRNA splicing. Chemical modulation of intron selection has emerged as a route for cancer therapy. Splicing modulators alter the splicing patterns in cells by binding to the U2 snRNP (small nuclear ribonucleoprotein)-a complex chaperoning the selection of branch and 3' splice sites. Here we report crystal structures of the SF3B module of the U2 snRNP in complex with spliceostatin and sudemycin FR901464 analogs, and the cryo-electron microscopy structure of a cross-exon prespliceosome-like complex arrested with spliceostatin A. The structures reveal how modulators inactivate the branch site in a sequence-dependent manner and stall an E-to-A prespliceosome intermediate by covalent coupling to a nucleophilic zinc finger belonging to the SF3B subunit PHF5A. These findings support a mechanism of intron recognition by the U2 snRNP as a toehold-mediated strand invasion and advance an unanticipated drug targeting concept. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7b92.cif.gz 7b92.cif.gz | 472.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7b92.ent.gz pdb7b92.ent.gz | 302.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7b92.json.gz 7b92.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b9/7b92 https://data.pdbj.org/pub/pdb/validation_reports/b9/7b92 ftp://data.pdbj.org/pub/pdb/validation_reports/b9/7b92 ftp://data.pdbj.org/pub/pdb/validation_reports/b9/7b92 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7b0iC  7b91C  7b9cC  7omfC  7onbC  7opiC  5ifeS  6en4S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
-Splicing factor 3B subunit ... , 3 types, 3 molecules ABC
| #1: Protein | Mass: 100722.109 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SF3B3, KIAA0017, SAP130 / Production host: Homo sapiens (human) / Gene: SF3B3, KIAA0017, SAP130 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: Q15393 Trichoplusia ni (cabbage looper) / References: UniProt: Q15393 |
|---|---|
| #2: Protein | Mass: 10149.369 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SF3B5, SF3B10 / Production host: Homo sapiens (human) / Gene: SF3B5, SF3B10 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: Q9BWJ5 Trichoplusia ni (cabbage looper) / References: UniProt: Q9BWJ5 |
| #3: Protein | Mass: 96615.734 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SF3B1, SAP155 / Production host: Homo sapiens (human) / Gene: SF3B1, SAP155 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: O75533 Trichoplusia ni (cabbage looper) / References: UniProt: O75533 |
-Protein , 1 types, 1 molecules D
| #4: Protein | Mass: 11670.525 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PHF5A / Production host: Homo sapiens (human) / Gene: PHF5A / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: Q7RTV0 Trichoplusia ni (cabbage looper) / References: UniProt: Q7RTV0 |
|---|
-Non-polymers , 3 types, 5 molecules 
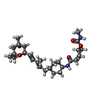



| #5: Chemical | | #6: Chemical | ChemComp-T2W / [(~{ | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.75 Å3/Da / Density % sol: 55.23 % / Description: Dome-shaped crystals |
|---|---|
| Crystal grow | Temperature: 277.15 K / Method: vapor diffusion, hanging drop Details: 0.1 M HEPES pH=7.42, 0.2 M Magnesium chloride, 27.75% (v/v) PEG-400 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 1 Å / Beamline: X10SA / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Feb 22, 2018 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3→49.06 Å / Num. obs: 49623 / % possible obs: 100 % / Redundancy: 20.8 % / Biso Wilson estimate: 106.3 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.181 / Rpim(I) all: 0.041 / Rrim(I) all: 0.185 / Net I/σ(I): 14.7 |
| Reflection shell | Resolution: 3→3.1 Å / Redundancy: 21.3 % / Rmerge(I) obs: 4.108 / Mean I/σ(I) obs: 0.8 / Num. unique obs: 4468 / CC1/2: 0.367 / Rpim(I) all: 0.907 / Rrim(I) all: 4.208 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5IFE, 6EN4 Resolution: 3→46.52 Å / SU ML: 0.6037 / Cross valid method: FREE R-VALUE / σ(F): 0.31 / Phase error: 31.3844 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 102.24 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3→46.52 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj



