+ Open data
Open data
- Basic information
Basic information
| Entry | 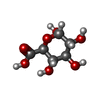 Database: PDB chemical components / ID: GCU Database: PDB chemical components / ID: GCU |
|---|---|
| Name | Name: Synonyms: alpha-D-glucuronic acid; D-glucuronic acid; glucuronic acid |
-Chemical information
| Composition |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Others | Type: D-saccharide, alpha linking / PDB classification: ATOMS / Three letter code: GCU / Model coordinates PDB-ID: 1DBO | ||||||||||||||||||
| History |
| ||||||||||||||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | [ | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | | OpenEye OEToolkits 1.5.0 | ( | |
|---|
-CONDENSED IUPAC CARBOHYDRATE SYMBOL
| GMML 1.0 |
|---|
-COMMON NAME
| GMML 1.0 |
|---|
-IUPAC CARBOHYDRATE SYMBOL
| PDB-CARE 1.0 |
|---|
-SNFG CARBOHYDRATE SYMBOL
| GMML 1.0 |
|---|
 Movie
Movie Controller
Controller