+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7944 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fako virus full particles, icosahedral reconstruction | |||||||||
 Map data Map data | An icosahedral reconstruction of Fako virus full particles | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | capsid / virion / Reoviridae / T=2 / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology information: / FAKV Clamp protein / : / : / : / : / Reovirus VP3 protein, guanylyltransferase (GTase) / Reovirus turret protein, bridge domain / Reovirus VP3 protein, Methyltransferase domain 1 / Reovirus VP3 protein, Methyltransferase domain 2 ...: / FAKV Clamp protein / : / : / : / : / Reovirus VP3 protein, guanylyltransferase (GTase) / Reovirus turret protein, bridge domain / Reovirus VP3 protein, Methyltransferase domain 1 / Reovirus VP3 protein, Methyltransferase domain 2 / : / Inner layer core protein VP1-like, C-terminal Similarity search - Domain/homology | |||||||||
| Biological species |   Fako virus Fako virus | |||||||||
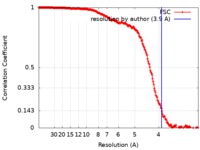
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Kaelber JT / Jiang W / Weaver SC / Auguste AJ / Chiu W | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Arrangement of the Polymerase Complexes inside a Nine-Segmented dsRNA Virus. Authors: Jason T Kaelber / Wen Jiang / Scott C Weaver / Albert J Auguste / Wah Chiu /  Abstract: Members of the family Reoviridae package several copies of the viral polymerase complex into their capsid to carry out replication and transcription within viral particles. Classical single-particle ...Members of the family Reoviridae package several copies of the viral polymerase complex into their capsid to carry out replication and transcription within viral particles. Classical single-particle reconstruction encounters difficulties resolving structures such as the intraparticle polymerase complex because refinement can converge to an incorrect map and because the map could depict a nonrepresentative subset of particles or an average of heterogeneous particles. Using the nine-segmented Fako virus, we tested hypotheses for the arrangement and number of polymerase complexes within the virion by measuring how well each hypothesis describes the set of cryoelectron microscopy images of individual viral particles. We find that the polymerase complex in Fako virus binds at ten possible sites despite having only nine genome segments. A single asymmetric configuration describes the arrangement of these complexes in both virions and genome-free capsids. Similarities between the arrangements of Reoviridae with 9, 10, and 11 segments indicate the generalizability of this architecture. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7944.map.gz emd_7944.map.gz | 685.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7944-v30.xml emd-7944-v30.xml emd-7944.xml emd-7944.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7944_fsc.xml emd_7944_fsc.xml | 20.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_7944.png emd_7944.png | 219.9 KB | ||
| Filedesc metadata |  emd-7944.cif.gz emd-7944.cif.gz | 7.3 KB | ||
| Others |  emd_7944_additional.map.gz emd_7944_additional.map.gz | 1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7944 http://ftp.pdbj.org/pub/emdb/structures/EMD-7944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7944 | HTTPS FTP |
-Related structure data
| Related structure data |  6djyMC  7941C  7945C  7948C  7949C  7953C  7954C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7944.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7944.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | An icosahedral reconstruction of Fako virus full particles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.506 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Another icosahedral reconstruction of Fako virus full particles
| File | emd_7944_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Another icosahedral reconstruction of Fako virus full particles | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Fako virus
| Entire | Name:   Fako virus Fako virus |
|---|---|
| Components |
|
-Supramolecule #1: Fako virus
| Supramolecule | Name: Fako virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1567404 / Sci species name: Fako virus / Sci species strain: CSW77 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Culicinae (mosquito) Culicinae (mosquito) |
| Molecular weight | Theoretical: 43 MDa |
| Virus shell | Shell ID: 1 / T number (triangulation number): 2 |
-Macromolecule #1: Clamp protein
| Macromolecule | Name: Clamp protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Molecular weight | Theoretical: 39.589691 KDa |
| Sequence | String: MTLTYWDKEK RMTLKQMIQQ VAINEQENEL THYVFTTPLS MPTFGKPMLG YVPLNEVATS KFFSNVNDFD RDNQLAMAHF PDTTITQAY NLTNSIKPGD TSLPDAEVAA LKWFWKFFTS INLVRQPPMD NVMYWACQFL SSGTSFLPLE RDVEIVFSGF K GSHICMFS ...String: MTLTYWDKEK RMTLKQMIQQ VAINEQENEL THYVFTTPLS MPTFGKPMLG YVPLNEVATS KFFSNVNDFD RDNQLAMAHF PDTTITQAY NLTNSIKPGD TSLPDAEVAA LKWFWKFFTS INLVRQPPMD NVMYWACQFL SSGTSFLPLE RDVEIVFSGF K GSHICMFS NLRQMNLSPI LCPYYDLITN FKTTTEIRAY VDAHEELKSL LTYLCLCTIV GLCDTFTETR NMDTGEYVWK VR DVVSRNH TPAQNVEKFC YTIQNAKYMI QLVHVLLFPL TDNKYADLPN YVAVITQGAI NQSRSHNVIN TTDESNSNTT SDT AASTSG IVSGDTGTVA SLYPDEFKYV QS UniProtKB: Clamp protein |
-Macromolecule #2: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Molecular weight | Theoretical: 136.689766 KDa |
| Sequence | String: MRPIRMYKNN QERTNLKHQE INEEQQNEQT TSNQGFTRSD NSGKINIERI SSSRNQITDG KTVSSYSKIE TNRSSQDSVQ HGGSSITYT SDTTGNPRIT NARTNNDETH ATGPIEDLNS TSHGREPEIE SFADRAELAM MIQGMTVGAL TVQPMRSIRS T FANLANVL ...String: MRPIRMYKNN QERTNLKHQE INEEQQNEQT TSNQGFTRSD NSGKINIERI SSSRNQITDG KTVSSYSKIE TNRSSQDSVQ HGGSSITYT SDTTGNPRIT NARTNNDETH ATGPIEDLNS TSHGREPEIE SFADRAELAM MIQGMTVGAL TVQPMRSIRS T FANLANVL IFHDVFTTED KPSAFIEYHS DEMIVNMPKQ TYNPIDNLAK ILYLPSLEKF KYGTGIVQLN YSPHISKLYQ NT NNIINTI TDGITYANRT EFFIRVMVLM MMDRKILTME FYDVDTSAIS NTAILPTIPT TTGVSPLLRI DTRTEPIWYN DAI KTLITN LTIQYGKIKT VLDANAVKRY SVVGYPIDQY RAYLYNHNLL EYLGKKVKRE DIMSLIKALS YEFDLITISD LEYQ NIPKW FSDNDLSRFI FSICMFPDIV RQFHALNIDY FSQANVFTVK SENAIVKMLN SNQNMEPTII NWFLFRICAI DKTVI DDYF SLEMTPIIMR PKLYDFDMKR GEPVSLLYIL ELILFSIMFP NVTQHMLGQI QARILYISMY AFRQEYLKFI TKFGFY YKI VNGRKEYIQV TNQNERMTEN NDVLTGNLYP SLFTDDPTLS AIAPTLAKIA RLMKPTTSLT PDDRAIAAKF PRFKDSA HL NPYSSLNIGG RTQHSVTYTR MYDAIEEMFN LILRAFASSF AQRPRAGVTQ LKSLLTQLAD PLCLALDGHV YHLYNVMA N MMQNFIPNTD GQFHSFRACS YAVKDGGNIY RVVQNGDELN ESLLIDTAIV WGLLGNTDSS YGNAIGATGT ANVPTKVQP VIPTPDNFIT PTIHLKTSID AICSVEGILL LILSRQTTIP GYEDELNKLR TGISQPKVTE RQYRRARESI KNMLGSGDYN VAPLHFLLH TEHRSTKLSK PLIRRVLDNV VQPYVANLDP AEFENTPQLI ENSNMTRLQI ALKMLTGDMD DIVKGLILHK R ACAKFDVY ETLTIPTDVK TIVLTMQHIS TQTQNNMVYY VFLIDGVKIL AEDIKNVNFQ IDITGIWPEY VITLLLRAIN NG FNTYVSM PNILYKPTIT ADVRQFMNTT KAETLLISNK SIVHEIMFFD NALQPKMSSD TLALSEAVYR TIWNSSIITQ RIS ARGLMN LEDARPPEAK ISHQSELDMG KIDETSGEPI YTSGLQKMQS SKVSMANVVL SAGSDVIRQA AIKYNVVRTQ EIIL FE UniProtKB: Major capsid protein |
-Macromolecule #3: Turret protein
| Macromolecule | Name: Turret protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Molecular weight | Theoretical: 121.23257 KDa |
| Sequence | String: MIDLRLEEDI LTATLPEFLS TRPKYRYAYT NTKQQDIRFQ GPMRHVRLTH LYKQTKLWNL QYIERELAIS EIDDALDEFI QTFSLPYVI EQGTYKYNML LGMHAHNVNY QDDVSELIAN NPQLLNYLDD NPFSAIFELV NVDLQIYQYG QNIFNNEAEH T ILFLKDNT ...String: MIDLRLEEDI LTATLPEFLS TRPKYRYAYT NTKQQDIRFQ GPMRHVRLTH LYKQTKLWNL QYIERELAIS EIDDALDEFI QTFSLPYVI EQGTYKYNML LGMHAHNVNY QDDVSELIAN NPQLLNYLDD NPFSAIFELV NVDLQIYQYG QNIFNNEAEH T ILFLKDNT NYGVIQALQK HPFSATHINW HLHKHIFVFH SREQLLNKLL SAGLEDSQLY QRQKTYSTKR GDRPTERMVT YI EDDHIRR IQAVFPLLLD NIFDVKLHKD SSMTWLKSYA DMIYDSVKNS NSTITPEIRK LYLRMYNQYM RIFLPIEQYM LYD NTCWPF SEKITLKINV RLISSRENQP VLWKTPIDTE NLISIVQPDE PINKLNFTAI PSTMIRLNDN ITMYRAVKDM FSAI EYLPD AIENIPTLTM KEQALSRYIS PDSEAQNFFN NQPPYLNSIM NVNRQVFEAV KRGNIQVSTG SMEHLCLCMH VKSGL IVGR TVLIDDKVVL RRNFNASTAK MITCYVKAFA QLYGEGSLIN PGLRMVFFGV ETEPAIDILK LFYGDKSLYI QGFGDR GIG RDKFRTKIED ALTLRIGCDI LISDIDQADY EDPNEEKFDD ITDFVCYVTE LVISNATVGL VKISMPTYYI MNKISST LN NKFSNVAINI VKLSTQKPYT YEAYIMLSHG STLTNKGYLR NPVCDVYLEK ISLQPMDLKI ISTISNEINY DKPTLYRF V VDKNDVTDVS IAMHILSIHC STITTRSVMV RSDNTGAFVT MSGIKDMKRV AIMNRMTDGT SANSYMHEQN GKLYLQKVP YLEDLISAFP NGFGSTYQND YDSSMSVINV NALIRQVVYR VISKSIPVAL LESLSRIRII GGRDLGEMNA VYKLYKTPIE VYDAVGITR EYPHVQISYR AQRYSFTESI PNHTLLLANY VIMNDVDGAP ISSLEQINTI KKIISKISLG SIAYIQVYTD I VARNINVM TKNDSFLISA NADKTVFKVQ VSGYKAVEMC NYEQLLQLVS DNTGVNIIKL TYQDVLESCV LSSGILGDTG SW LLDLVLA STYIIEIRG UniProtKB: Turret protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Details: unspecified | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Number real images: 2400 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.7 mm |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)