[English] 日本語
 Yorodumi
Yorodumi- EMDB-6904: Cryo-EM structure of human Dicer and its complexes with a pre-miR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6904 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | |||||||||
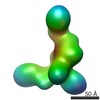
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dicer / TRBP / Cryo-EM / RNA interference / HYDROLASE-PROTEIN BINDING complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of siRNA processing / regulation of miRNA processing / negative regulation of cytoplasmic pattern recognition receptor signaling pathway / regulation of viral transcription / peripheral nervous system myelin formation / regulation of regulatory ncRNA processing / negative regulation of defense response to virus by host / global gene silencing by mRNA cleavage / pre-miRNA binding / negative regulation of Schwann cell proliferation ...regulation of siRNA processing / regulation of miRNA processing / negative regulation of cytoplasmic pattern recognition receptor signaling pathway / regulation of viral transcription / peripheral nervous system myelin formation / regulation of regulatory ncRNA processing / negative regulation of defense response to virus by host / global gene silencing by mRNA cleavage / pre-miRNA binding / negative regulation of Schwann cell proliferation / tRNA-derived small RNA (tsRNA or tRNA-related fragment, tRF) biogenesis / Small interfering RNA (siRNA) biogenesis / apoptotic DNA fragmentation / tRNA decay / ribonuclease III / deoxyribonuclease I activity / positive regulation of Schwann cell differentiation / nerve development / RISC-loading complex / positive regulation of myelination / miRNA metabolic process / RISC complex assembly / miRNA processing / ribonuclease III activity / skeletal muscle tissue regeneration / pre-miRNA processing / siRNA processing / siRNA binding / M-decay: degradation of maternal mRNAs by maternally stored factors / Regulation of MITF-M-dependent genes involved in apoptosis / pre-mRNA binding / RISC complex / miRNA binding / neural precursor cell proliferation / MicroRNA (miRNA) biogenesis / positive regulation of muscle cell differentiation / negative regulation of tumor necrosis factor production / spermatid development / negative regulation of tumor necrosis factor-mediated signaling pathway / single fertilization / positive regulation of viral genome replication / negative regulation of protein kinase activity / neuron projection morphogenesis / RNA endonuclease activity / protein sequestering activity / positive regulation of translation / helicase activity / PKR-mediated signaling / multicellular organism growth / double-stranded RNA binding / nuclear body / protein domain specific binding / negative regulation of gene expression / perinuclear region of cytoplasm / enzyme binding / negative regulation of transcription by RNA polymerase II / protein homodimerization activity / RNA binding / extracellular exosome / nucleoplasm / ATP binding / metal ion binding / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Liu Z / Wang J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Authors: Zhongmin Liu / Jia Wang / Hang Cheng / Xin Ke / Lei Sun / Qiangfeng Cliff Zhang / Hong-Wei Wang /  Abstract: Human Dicer (hDicer) is a multi-domain protein belonging to the RNase III family. It plays pivotal roles in small RNA biogenesis during the RNA interference (RNAi) pathway by processing a diverse ...Human Dicer (hDicer) is a multi-domain protein belonging to the RNase III family. It plays pivotal roles in small RNA biogenesis during the RNA interference (RNAi) pathway by processing a diverse range of double-stranded RNA (dsRNA) precursors to generate ∼22 nt microRNA (miRNA) or small interfering RNA (siRNA) products for sequence-directed gene silencing. In this work, we solved the cryoelectron microscopy (cryo-EM) structure of hDicer in complex with its cofactor protein TRBP and revealed the precise spatial arrangement of hDicer's multiple domains. We further solved structures of the hDicer-TRBP complex bound with pre-let-7 RNA in two distinct conformations. In combination with biochemical analysis, these structures reveal a property of the hDicer-TRBP complex to promote the stability of pre-miRNA's stem duplex in a pre-dicing state. These results provide insights into the mechanism of RNA processing by hDicer and illustrate the regulatory role of hDicer's N-terminal helicase domain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6904.map.gz emd_6904.map.gz | 2.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6904-v30.xml emd-6904-v30.xml emd-6904.xml emd-6904.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6904.png emd_6904.png | 47.1 KB | ||
| Filedesc metadata |  emd-6904.cif.gz emd-6904.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6904 http://ftp.pdbj.org/pub/emdb/structures/EMD-6904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6904 | HTTPS FTP |
-Validation report
| Summary document |  emd_6904_validation.pdf.gz emd_6904_validation.pdf.gz | 383.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6904_full_validation.pdf.gz emd_6904_full_validation.pdf.gz | 382.6 KB | Display | |
| Data in XML |  emd_6904_validation.xml.gz emd_6904_validation.xml.gz | 5.5 KB | Display | |
| Data in CIF |  emd_6904_validation.cif.gz emd_6904_validation.cif.gz | 6.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6904 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6904 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6904 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6904 | HTTPS FTP |
-Related structure data
| Related structure data |  5zakMC  6905C  6906C  5zalC  5zamC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6904.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6904.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.30654 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
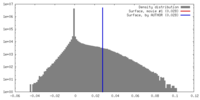
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dicer and trbp
| Entire | Name: Dicer and trbp |
|---|---|
| Components |
|
-Supramolecule #1: Dicer and trbp
| Supramolecule | Name: Dicer and trbp / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 250 KDa |
-Supramolecule #2: Human Dicer and TRBP complex
| Supramolecule | Name: Human Dicer and TRBP complex / type: complex / ID: 2 / Parent: 1 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Endoribonuclease Dicer
| Macromolecule | Name: Endoribonuclease Dicer / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ribonuclease III |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 218.947328 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKSPALQPLS MAGLQLMTPA SSPMGPFFGL PWQQEAIHDN IYTPRKYQVE LLEAALDHNT IVCLNTGSGK TFIAVLLTKE LSYQIRGDF SRNGKRTVFL VNSANQVAQQ VSAVRTHSDL KVGEYSNLEV NASWTKERWN QEFTKHQVLI MTCYVALNVL K NGYLSLSD ...String: MKSPALQPLS MAGLQLMTPA SSPMGPFFGL PWQQEAIHDN IYTPRKYQVE LLEAALDHNT IVCLNTGSGK TFIAVLLTKE LSYQIRGDF SRNGKRTVFL VNSANQVAQQ VSAVRTHSDL KVGEYSNLEV NASWTKERWN QEFTKHQVLI MTCYVALNVL K NGYLSLSD INLLVFDECH LAILDHPYRE IMKLCENCPS CPRILGLTAS ILNGKCDPEE LEEKIQKLEK ILKSNAETAT DL VVLDRYT SQPCEIVVDC GPFTDRSGLY ERLLMELEEA LNFINDCNIS VHSKERDSTL ISKQILSDCR AVLVVLGPWC ADK VAGMMV RELQKYIKHE QEELHRKFLL FTDTFLRKIH ALCEEHFSPA SLDLKFVTPK VIKLLEILRK YKPYERQQFE SVEW YNNRN QDNYVSWSDS EDDDEDEEIE EKEKPETNFP SPFTNILCGI IFVERRYTAV VLNRLIKEAG KQDPELAYIS SNFIT GHGI GKNQPRNKQM EAEFRKQEEV LRKFRAHETN LLIATSIVEE GVDIPKCNLV VRFDLPTEYR SYVQSKGRAR APISNY IML ADTDKIKSFE EDLKTYKAIE KILRNKCSKS VDTGETDIDP VMDDDDVFPP YVLRPDDGGP RVTINTAIGH INRYCAR LP SDPFTHLAPK CRTRELPDGT FYSTLYLPIN SPLRASIVGP PMSCVRLAER VVALICCEKL HKIGELDDHL MPVGKETV K YEEELDLHDE EETSVPGRPG STKRRQCYPK AIPECLRDSY PRPDQPCYLY VIGMVLTTPL PDELNFRRRK LYPPEDTTR CFGILTAKPI PQIPHFPVYT RSGEVTISIE LKKSGFMLSL QMLELITRLH QYIFSHILRL EKPALEFKPT DADSAYCVLP LNVVNDSST LDIDFKFMED IEKSEARIGI PSTKYTKETP FVFKLEDYQD AVIIPRYRNF DQPHRFYVAD VYTDLTPLSK F PSPEYETF AEYYKTKYNL DLTNLNQPLL DVDHTSSRLN LLTPRHLNQK GKALPLSSAE KRKAKWESLQ NKQILVPELC AI HPIPASL WRKAVCLPSI LYRLHCLLTA EELRAQTASD AGVGVRSLPA DFRYPNLDFG WKKSIDSKSF ISISNSSSAE NDN YCKHST IVPENAAHQG ANRTSSLENH DQMSVNCRTL LSESPGKLHV EVSADLTAIN GLSYNQNLAN GSYDLANRDF CQGN QLNYY KQEIPVQPTT SYSIQNLYSY ENQPQPSDEC TLLSNKYLDG NANKSTSDGS PVMAVMPGTT DTIQVLKGRM DSEQS PSIG YSSRTLGPNP GLILQALTLS NASDGFNLER LEMLGDSFLK HAITTYLFCT YPDAHEGRLS YMRSKKVSNC NLYRLG KKK GLPSRMVVSI FDPPVNWLPP GYVVNQDKSN TDKWEKDEMT KDCMLANGKL DEDYEEEDEE EESLMWRAPK EEADYED DF LEYDQEHIRF IDNMLMGSGA FVKKISLSPF STTDSAYEWK MPKKSSLGSM PFSSDFEDFD YSSWDAMCYL DPSKAVEE D DFVVGFWNPS EENCGVDTGK QSISYDLHTE QCIADKSIAD CVEALLGCYL TSCGERAAQL FLCSLGLKVL PVIKRTDRE KALCPTRENF NSQQKNLSVS CAAASVASSR SSVLKDSEYG CLKIPPRCMF DHPDADKTLN HLISGFENFE KKINYRFKNK AYLLQAFTH ASYHYNTITD CYQRLEFLGD AILDYLITKH LYEDPRQHSP GVLTDLRSAL VNNTIFASLA VKYDYHKYFK A VSPELFHV IDDFVQFQLE KNEMQGMDSE LRRSEEDEEK EEDIEVPKAM GDIFESLAGA IYMDSGMSLE TVWQVYYPMM RP LIEKFSA NVPRSPVREL LEMEPETAKF SPAERTYDGK VRVTVEVVGK GKFKGVGRSY RIAKSAAARR ALRSLKANQP QVP NS UniProtKB: Endoribonuclease Dicer |
-Macromolecule #2: RISC-loading complex subunit TARBP2
| Macromolecule | Name: RISC-loading complex subunit TARBP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.085277 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSEEEQGSGT TTGCGLPSIE QMLAANPGKT PISLLQEYGT RIGKTPVYDL LKAEGQAHQP NFTFRVTVGD TSCTGQGPSK KAAKHKAAE VALKHLKGGS MLEPALEDSS SFSPLDSSLP EDIPVFTAAA AATPVPSVVL TRSPPMELQP PVSPQQSECN P VGALQELV ...String: MSEEEQGSGT TTGCGLPSIE QMLAANPGKT PISLLQEYGT RIGKTPVYDL LKAEGQAHQP NFTFRVTVGD TSCTGQGPSK KAAKHKAAE VALKHLKGGS MLEPALEDSS SFSPLDSSLP EDIPVFTAAA AATPVPSVVL TRSPPMELQP PVSPQQSECN P VGALQELV VQKGWRLPEY TVTQESGPAH RKEFTMTCRV ERFIEIGSGT SKKLAKRNAA AKMLLRVHTV PLDARDGNEV EP DDDHFSI GVGSRLDGLR NRGPGCTWDS LRNSVGEKIL SLRSCSLGSL GALGPACCRV LSELSEEQAF HVSYLDIEEL SLS GLCQCL VELSTQPATV CHGSATTREA ARGEAARRAL QYLKIMAGSK UniProtKB: RISC-loading complex subunit TARBP2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, 2 mM EDTA |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Details: The grid was glow-discharged prior to use. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was mono-disperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 80.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-32 / Number grids imaged: 1 / Number real images: 7581 / Average exposure time: 8.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.5 µm / Calibrated defocus min: 2.5 µm / Calibrated magnification: 38462 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 100 / Target criteria: CC | ||||||||||
| Output model |  PDB-5zak: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)