[English] 日本語
 Yorodumi
Yorodumi- EMDB-51017: Structure of the Partially-assembled gamma-Tubulin Ring Complex f... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Partially-assembled gamma-Tubulin Ring Complex from Pig Brain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tubulin Complex / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgamma-tubulin complex localization / microtubule nucleator activity / Recruitment of mitotic centrosome proteins and complexes / polar microtubule / gamma-tubulin complex / gamma-tubulin ring complex / meiotic spindle organization / microtubule nucleation / gamma-tubulin binding / Recruitment of NuMA to mitotic centrosomes ...gamma-tubulin complex localization / microtubule nucleator activity / Recruitment of mitotic centrosome proteins and complexes / polar microtubule / gamma-tubulin complex / gamma-tubulin ring complex / meiotic spindle organization / microtubule nucleation / gamma-tubulin binding / Recruitment of NuMA to mitotic centrosomes / pericentriolar material / mitotic sister chromatid segregation / cytoplasmic microtubule / spindle assembly / cytoplasmic microtubule organization / centriole / mitotic spindle organization / spindle microtubule / meiotic cell cycle / brain development / spindle / neuron migration / spindle pole / mitotic cell cycle / microtubule binding / microtubule / ciliary basal body / centrosome / GTP binding / nucleoplasm / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
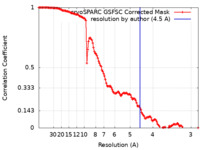
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Munoz-Hernandez H / Wieczorek M | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Dev Cell / Year: 2024 Journal: Dev Cell / Year: 2024Title: Partial closure of the γ-tubulin ring complex by CDK5RAP2 activates microtubule nucleation. Authors: Yixin Xu / Hugo Muñoz-Hernández / Rościsław Krutyhołowa / Florina Marxer / Ferdane Cetin / Michal Wieczorek /  Abstract: Microtubule nucleation is templated by the γ-tubulin ring complex (γ-TuRC), but its structure deviates from the geometry of α-/β-tubulin in the microtubule, explaining the complex's poor ...Microtubule nucleation is templated by the γ-tubulin ring complex (γ-TuRC), but its structure deviates from the geometry of α-/β-tubulin in the microtubule, explaining the complex's poor nucleating activity. Several proteins may activate the γ-TuRC, but the mechanisms underlying activation are not known. Here, we determined the structure of the porcine γ-TuRC purified using CDK5RAP2's centrosomin motif 1 (CM1). We identified an unexpected conformation of the γ-TuRC bound to multiple protein modules containing MZT2, GCP2, and CDK5RAP2, resulting in a long-range constriction of the γ-tubulin ring that brings it in closer agreement with the 13-protofilament microtubule. Additional CDK5RAP2 promoted γ-TuRC decoration and stimulated the microtubule-nucleating activities of the porcine γ-TuRC and a reconstituted, CM1-free human complex in single-molecule assays. Our results provide a structural mechanism for the control of microtubule nucleation by CM1 proteins and identify conformational transitions in the γ-TuRC that prime it for microtubule nucleation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51017.map.gz emd_51017.map.gz | 108.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51017-v30.xml emd-51017-v30.xml emd-51017.xml emd-51017.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51017_fsc.xml emd_51017_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_51017.png emd_51017.png | 71 KB | ||
| Filedesc metadata |  emd-51017.cif.gz emd-51017.cif.gz | 8 KB | ||
| Others |  emd_51017_half_map_1.map.gz emd_51017_half_map_1.map.gz emd_51017_half_map_2.map.gz emd_51017_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51017 http://ftp.pdbj.org/pub/emdb/structures/EMD-51017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51017 | HTTPS FTP |
-Validation report
| Summary document |  emd_51017_validation.pdf.gz emd_51017_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_51017_full_validation.pdf.gz emd_51017_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_51017_validation.xml.gz emd_51017_validation.xml.gz | 21.9 KB | Display | |
| Data in CIF |  emd_51017_validation.cif.gz emd_51017_validation.cif.gz | 28.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51017 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51017 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51017 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51017 | HTTPS FTP |
-Related structure data
| Related structure data |  9g3xMC  9g3yC  9g3zC  9g40C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_51017.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51017.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4133 Å | ||||||||||||||||||||||||||||||||||||
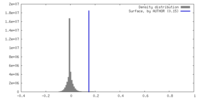
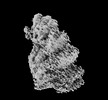
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_51017_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
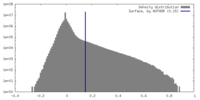
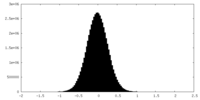
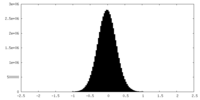
| Density Histograms |
-Half map: #2
| File | emd_51017_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
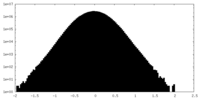
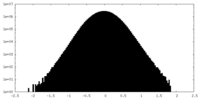
| Density Histograms |
- Sample components
Sample components
-Entire : Gamma-Tubulin Ring Complex in native pig brain
| Entire | Name: Gamma-Tubulin Ring Complex in native pig brain |
|---|---|
| Components |
|
-Supramolecule #1: Gamma-Tubulin Ring Complex in native pig brain
| Supramolecule | Name: Gamma-Tubulin Ring Complex in native pig brain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mitotic spindle organizing protein 1
| Macromolecule | Name: Mitotic spindle organizing protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.285489 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGNSGSGAA AAANLNAVRE TMDVLLEISR ILNTGLDMET LSICVRLCEQ GINPEALSSV IKELRKATEA LKAAENMTS UniProtKB: Mitotic spindle organizing protein 1 |
-Macromolecule #2: Gamma-tubulin complex component
| Macromolecule | Name: Gamma-tubulin complex component / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 102.609703 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSEFRIHHDV NELLSLLRVH GGDGAEVYID LLQKNRTPYV TTTVSAHSAK VKIAEFSRTP EDFLKKYDEL KSKNTRNLDP LVYLLSKLM EDRETLQYLQ QNAKERAELA ASAAASSTAS FGASATASKI SMQELEELRK QLGSVATGPT WQQSLELTRK M LRDKQSKK ...String: MSEFRIHHDV NELLSLLRVH GGDGAEVYID LLQKNRTPYV TTTVSAHSAK VKIAEFSRTP EDFLKKYDEL KSKNTRNLDP LVYLLSKLM EDRETLQYLQ QNAKERAELA ASAAASSTAS FGASATASKI SMQELEELRK QLGSVATGPT WQQSLELTRK M LRDKQSKK NSGQRLPVLP AWVYERPALL GDFLPGTGGS ADTAVPIGSL PLASQEAAVV EDLLYVLVGV DGRYISAQPL TG RQGRTFL VDPNLDLSIR ELVSRILPVA ASYSTVTRFI EEKSSFEYGQ VNHALAAAMR TLVKEYLVLV TQLEQLQRQG LLS LQKLWF YIQPAMRSLD ILASLATSVD KGECIGGATL SLLHDRSFSY TGDSQAQELC LHLTKAASTP YFEILEKWIY RGII DDPYS EFMVEEHELR KEKIQEDYND KYWDQRYTVV QRQIPSFLQK MAGKVLSTGK YLNVVRECGH DVTCPVAKEV VYTLK ERAY VEQIEKAFSY ASKVLLDFLM GEKELLAHLR SIKRYFLMDQ GDFFVHFMDL TEEELKKPVD DITPTRLEAL LELALR MST ANTDPFKDDL KIDLMPHDLI TQLLRVLAIE TQQEKAMVHA DPTELTLSGL EAFSFDYVVT WPLSLIINRK ALTRYQM LF RHMFYCKHVE RQLCSVWISN KAAKRFSLHS AKWFAGAFTL RQRMLNFVQN IQSYMMFEVM EPTWHVLEQN LRSASNID D VLGHHASFLD NCLKDCMLTN PELLRVFSKL MSVCVMFTNC LQRFTQSMKL DSELGHPALE PGAMLGPPTE AERAEERAR KELARKCLAE HVDAPQLASS FEATITKFDK NFSAHLLDLL ARLSIYSTSD CEHGMASVIS RLDFNGFYTE RLERLSAERS QKAAPPVPG PRGPPALVPR VAVTAQ UniProtKB: Gamma-tubulin complex component |
-Macromolecule #3: Gamma-tubulin complex component 3
| Macromolecule | Name: Gamma-tubulin complex component 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 103.172477 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATPDQKSPN VLLQNLCCRI LGKSEADVAQ QFQYAVRVIG SNFAPTVERD EFLVAEKIKK ELTRQRREAD AALFSELHRK LHSQGVLKN KWSILYLLLS LSEDPRKQPS KVSGYAALFA QALPRDAHST PYYYARPQSL PLNYQERGAP SAQSAGSAGS S GVSSLGTY ...String: MATPDQKSPN VLLQNLCCRI LGKSEADVAQ QFQYAVRVIG SNFAPTVERD EFLVAEKIKK ELTRQRREAD AALFSELHRK LHSQGVLKN KWSILYLLLS LSEDPRKQPS KVSGYAALFA QALPRDAHST PYYYARPQSL PLNYQERGAP SAQSAGSAGS S GVSSLGTY ALNGPTPPPP PPALLPGQPL PAPGVGDGLR QQLGSRLAWT LTASQPSLPS TTSKAVPSSG SRGAARPRRE GD AAAGAVE VTEAALVRDI LYVFQGIDGK HVKMSNADNC YTVEGKANLS KSLRDTAVRL AELGWLHNKI RKYTDQRSLD RSF GLVGQS FCAALHQELR EYYRLLSVLH SQLQLEDDQG VNLGLESSLT LRRLLVWTYD PKMRLKTLAA LVDHCQGRKG GELA SAVHA YTKTGDPYAR SLVQHILSLV SHPVLSFLYR WIYDGELEDT YHEFFVASDP AVKADRLWHD KYALRKPMIP SFMTM DQCR KVLLIGKSIN FLHQVCHDQT PTTKMIAVTK SAESPQDAAD LFTDLENAFQ GKIDAAYFET SKYLLDVLNK KYSLLD HMQ AMRRYLLLGQ GDFIRHLMDL LKPELVRPAT TLYQHNLTGI LETAVRATNA QFDSPEILKR LDVRLLEVSP GDTGWDV FS LDYHVDGPIA TVFTRECMSH YLRAFNFLWR AKRVEYILTD IRKGHMCNAR LLRSMPEFSG VLHHCHILAS EMVHFIHQ M QYYVTFEVLE CSWDELWNRV QRAQDLDHII AAHEAFLGTV ISRCLLDSDS RALLNQLRAV FDQIIELQNT QDAIYRAAL EELQRRLQFE EKKKQREAEG QWGVSAAEEE QEKRRVQEFQ ESIPKMCSQL RILTHFYQGV VQQFLVSLTT SSDESLRFLS FRLDFNEHY RAREPRLRVS LGTRGRRSSH T UniProtKB: Gamma-tubulin complex component 3 |
-Macromolecule #4: Gamma-tubulin complex component
| Macromolecule | Name: Gamma-tubulin complex component / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 76.104867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIHELLLALS GYPGSIFTWN KRGGLQVSQD FPFLHPSETS VLNRLCRLGT DYIRFTEFIE QYTGHVQQQD HHPSQQGQGG LHGIYLRAF CTGLDSVLQP YRQALLDLEQ EFLADPHLSI SHINYSLDQF QLLFPSVMVV VEQIKSQKIH GCQILETVYK H SCGGLPPV ...String: MIHELLLALS GYPGSIFTWN KRGGLQVSQD FPFLHPSETS VLNRLCRLGT DYIRFTEFIE QYTGHVQQQD HHPSQQGQGG LHGIYLRAF CTGLDSVLQP YRQALLDLEQ EFLADPHLSI SHINYSLDQF QLLFPSVMVV VEQIKSQKIH GCQILETVYK H SCGGLPPV RSALEKILAV CHGVMYKQLS AWMLHGLLLD QHEEFFIKQG PSSGNVSAQP EEDEEDLGIG GLTGKQLREL QD LRLIEEE NMLAPSLKQF SLRVEILPSY IPVRVAEKIL FVGESVQMFE NQNVNLTRKG SILKDQEDTF AAELHRLKQQ PLF SLVDFE QVVDRIRSTV AEHLWKLMVE ESDLLGQLKI IKDFYLLGRG ELFQAFIDTA QHMLKTPPTA VTEHDVNVAF QQSA HKVLL DDDNLLPLLH LTIEYHGKEH KADATQAREG PSRETSPREA PASGWAALGL SYKVQWPLHI LFTPAVLEKY NVVFK YLLS VRRVQAELQH CWALQMQRKH LKSNQTDAVK WRLRNHMAFL VDNLQYYLQV DVLESQFSQL LHQINSTRDF ESIRLA HDH FLSNLLAQSF ILLKPVFHCL NEILDLCHSF CSLVSQNLGP LDERGAAQLS ILVKGFSRQS SLLFKILSSV RNHQINS DL AQLLLRLDYN KYYTQAGGTL GSFGM UniProtKB: Gamma-tubulin complex component |
-Macromolecule #5: Gamma-tubulin complex component
| Macromolecule | Name: Gamma-tubulin complex component / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 122.040344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASPAPSWTR LDPQQERDVR ELIRLVSGVQ DEADPNFQLA LHFAWSNFRF HRFLDVNSHK VEKTIEGIYE KFIIHSDLSK AASWKRLTD EFLNASLPSI KEIKTDAHYS ILSLLLCLSD SPSNSNYVET PRNKEVEKKD DFDWGKYLME GEEIDLGPNV D TPNWSEES ...String: MASPAPSWTR LDPQQERDVR ELIRLVSGVQ DEADPNFQLA LHFAWSNFRF HRFLDVNSHK VEKTIEGIYE KFIIHSDLSK AASWKRLTD EFLNASLPSI KEIKTDAHYS ILSLLLCLSD SPSNSNYVET PRNKEVEKKD DFDWGKYLME GEEIDLGPNV D TPNWSEES EDEDDPQPLS REDSGIQVDR TPLEEQDQSR KPASRVSWKV DEPDARSWLE QHVVRQYWTT RSSKFPHSLH LH SNLAAVW DQHLYSSDPL YVPDDRVSVT ETQVIRETLW LLSGVKKLFI FQLIDGKVAV RNNIMVTHLT HSCLRSVLEQ IAA YGQVVF RLQEFIDEVM GHSSESTLPG NGSVPKKSTD APFRTYQAFM WALYKYFISF KEELSEIEKC IINNDTTVTL AIVV DKLSP RLAQLKVLHK VFSTGVAEVP PDTRNVVRAS HLLNTLYKAI LEYDNVGEAS EQTVSLLFSL WVETVRPYLQ IVDEW IVHG HLCDGAREFI IQRNKNVPVN HRDFWYATYT LYSVSEKTEN EEKMSDNASA SSGSDQGPSS RQHTMVSFLK PVLKQI IMA GKSMQLLKNL QCAESTTCQA MARDAERKSL YTLFLESVQS RLRHGEDATA QALTEQQATR ETLIKMQSIA ERHLELD DV HDPLLAINFA RLYLEQSDFH EKFAGGDICV DRSSESVTCQ TFELTLRSCL YPHIDKQYLD CCGNLMRTLK KDYRLVEY L QAMRNFFLME GGDTMYDFYT SIFDKIREKE TWQNVSFLNV QLQEAVGQRY PEDSSRLSIS FENTDTAKKK LPVHTLDGL TLSYKVPWPV DIVISLECQK IYNQVFLLLL QIKWAKYSLD VLLFGELASS AEKPQSKEGL LSGQDTAAQF GPQKEPVRQQ IHRMFLLRV KLMHFVNSLH NYIMTRILHS TGLEFQHQVE EAKDLDQLIK IHYRYLSTIH DRCLLREKVS FVKEAIMKVL N LALMFADG WQAGLGAWQM ESIEKMESDF KNCHMFLVTI LNKAVCRGSF PHLESLALSL MAGMEQKRED DLFNHTWGQG DL PNYTCAV RLLGVRKGGG TRPK UniProtKB: Gamma-tubulin complex component |
-Macromolecule #6: Tubulin gamma chain
| Macromolecule | Name: Tubulin gamma chain / type: protein_or_peptide / ID: 6 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.135562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPREIITLQL GQCGNQIGFE FWKQLCAEHG ISPEGIVEEF ATEGTDRKDV FFYQADDEHY IPRAVLLDLE PRVIHSILNS PYAKLYNPE NIYLSEHGGG AGNNWASGFS QGEKIHEDIF DIIDREADGS DSLEGFVLCH SIAGGTGSGL GSYLLERLND R YPKKLVQT ...String: MPREIITLQL GQCGNQIGFE FWKQLCAEHG ISPEGIVEEF ATEGTDRKDV FFYQADDEHY IPRAVLLDLE PRVIHSILNS PYAKLYNPE NIYLSEHGGG AGNNWASGFS QGEKIHEDIF DIIDREADGS DSLEGFVLCH SIAGGTGSGL GSYLLERLND R YPKKLVQT YSVFPNQDEM SDVVVQPYNS LLTLKRLTQN ADCVVVLDNT ALNRIATDRL HIQNPSFSQI NQLVSTIMSA ST TTLRYPG YMNNDLIGLI ASLIPTPRLH FLMTGYTPLT TDQSVASVRK TTVLDVMRRL LQPKNVMVST GRDRQTNHCY IAI LNIIQG EVDPTQVHKS LQRIRERKLA NFIPWGPASI QVALSRKSPY LPSAHRVSGL MMANHTSISS LFESSCQQYD KLRK REAFL EQFRKEDIFK ENFDELDRSR EVVQELIDEY HAATRPDYIS WGTQEQ UniProtKB: Tubulin gamma chain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-9g3x: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)