+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of C-terminal LRRK2 bound to MLi-2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GTPase / kinase / inhibitors / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationcaveola neck / : / beta-catenin destruction complex binding / regulation of branching morphogenesis of a nerve / Wnt signalosome assembly / regulation of kidney size / regulation of cell projection organization / tangential migration from the subventricular zone to the olfactory bulb / GTP-dependent protein kinase activity / regulation of SNARE complex assembly ...caveola neck / : / beta-catenin destruction complex binding / regulation of branching morphogenesis of a nerve / Wnt signalosome assembly / regulation of kidney size / regulation of cell projection organization / tangential migration from the subventricular zone to the olfactory bulb / GTP-dependent protein kinase activity / regulation of SNARE complex assembly / regulation of neuroblast proliferation / regulation of ER to Golgi vesicle-mediated transport / protein localization to endoplasmic reticulum exit site / peroxidase inhibitor activity / negative regulation of late endosome to lysosome transport / regulation of mitochondrial depolarization / : / positive regulation of dopamine receptor signaling pathway / amphisome / regulation of synaptic vesicle transport / regulation of lysosomal lumen pH / regulation of CAMKK-AMPK signaling cascade / co-receptor binding / negative regulation of GTPase activity / regulation of dopamine receptor signaling pathway / positive regulation of microglial cell activation / regulation of neuron maturation / regulation of retrograde transport, endosome to Golgi / positive regulation of synaptic vesicle endocytosis / cytoplasmic side of mitochondrial outer membrane / negative regulation of autophagosome assembly / olfactory bulb development / JUN kinase kinase kinase activity / regulation of cAMP/PKA signal transduction / neuron projection arborization / striatum development / multivesicular body, internal vesicle / negative regulation of excitatory postsynaptic potential / regulation of dendritic spine morphogenesis / mitochondrion localization / protein localization to mitochondrion / cellular response to dopamine / positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / endoplasmic reticulum organization / positive regulation of protein autoubiquitination / negative regulation of protein processing / Wnt signalosome / positive regulation of programmed cell death / GTP metabolic process / regulation of canonical Wnt signaling pathway / syntaxin-1 binding / regulation of reactive oxygen species metabolic process / lysosome organization / Golgi-associated vesicle / clathrin binding / PTK6 promotes HIF1A stabilization / negative regulation of macroautophagy / regulation of mitochondrial fission / regulation of locomotion / protein kinase A binding / neuromuscular junction development / regulation of synaptic vesicle exocytosis / intracellular distribution of mitochondria / Golgi organization / microvillus / exploration behavior / endoplasmic reticulum exit site / autolysosome / locomotory exploration behavior / negative regulation of Notch signaling pathway / MAP kinase kinase kinase activity / canonical Wnt signaling pathway / regulation of synaptic vesicle endocytosis / regulation of synaptic transmission, glutamatergic / negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway / Rho protein signal transduction / presynaptic cytosol / neuron projection morphogenesis / phagocytic vesicle / cellular response to manganese ion / JNK cascade / positive regulation of autophagy / dendrite cytoplasm / GTPase activator activity / tubulin binding / cellular response to starvation / positive regulation of protein ubiquitination / SNARE binding / determination of adult lifespan / mitochondrion organization / cellular response to reactive oxygen species / excitatory postsynaptic potential / regulation of membrane potential / trans-Golgi network / calcium-mediated signaling / regulation of protein stability / regulation of autophagy / autophagy / small GTPase binding / mitochondrial membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
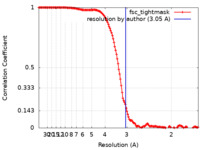
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Sanz-Murillo M / Villagran-Suarez A / Alegrio-Louro J / Leschziner A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures. Authors: Marta Sanz Murillo / Amalia Villagran Suarez / Verena Dederer / Deep Chatterjee / Jaime Alegrio Louro / Stefan Knapp / Sebastian Mathea / Andres E Leschziner /   Abstract: Mutations in leucine-rich repeat kinase 2 (LRRK2) are a common cause of familial Parkinson's disease (PD) and a risk factor for the sporadic form. Increased kinase activity was shown in patients with ...Mutations in leucine-rich repeat kinase 2 (LRRK2) are a common cause of familial Parkinson's disease (PD) and a risk factor for the sporadic form. Increased kinase activity was shown in patients with both familial and sporadic PD, making LRRK2 kinase inhibitors a major focus of drug development efforts. Although much progress has been made in understanding the structural biology of LRRK2, there are no available structures of LRRK2 inhibitor complexes. To this end, we solved cryo-electron microscopy structures of LRRK2, wild-type and PD-linked mutants, bound to the LRRK2-specific type I inhibitor MLi-2 and the broad-spectrum type II inhibitor GZD-824. Our structures revealed an active-like LRRK2 kinase in the type I inhibitor complex, and an inactive DYG-out in the type II inhibitor complex. Our structural analysis also showed how inhibitor-induced conformational changes in LRRK2 are affected by its autoinhibitory N-terminal repeats. The structures provide a template for the rational development of LRRK2 kinase inhibitors covering both canonical inhibitor binding modes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41709.map.gz emd_41709.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41709-v30.xml emd-41709-v30.xml emd-41709.xml emd-41709.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41709_fsc.xml emd_41709_fsc.xml | 15 KB | Display |  FSC data file FSC data file |
| Images |  emd_41709.png emd_41709.png | 56.6 KB | ||
| Filedesc metadata |  emd-41709.cif.gz emd-41709.cif.gz | 7.2 KB | ||
| Others |  emd_41709_additional_1.map.gz emd_41709_additional_1.map.gz emd_41709_half_map_1.map.gz emd_41709_half_map_1.map.gz emd_41709_half_map_2.map.gz emd_41709_half_map_2.map.gz | 4.5 MB 226.6 MB 226.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41709 http://ftp.pdbj.org/pub/emdb/structures/EMD-41709 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41709 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41709 | HTTPS FTP |
-Related structure data
| Related structure data |  8txzMC  8tyqC  8tzbC  8tzcC  8tzeC  8tzfC  8tzgC  8tzhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41709.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41709.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
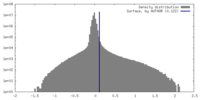
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_41709_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_41709_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
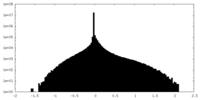
| Density Histograms |
-Half map: #1
| File | emd_41709_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
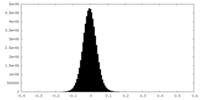
| Density Histograms |
- Sample components
Sample components
-Entire : C-terminal LRRK2 bound to MLi-2
| Entire | Name: C-terminal LRRK2 bound to MLi-2 |
|---|---|
| Components |
|
-Supramolecule #1: C-terminal LRRK2 bound to MLi-2
| Supramolecule | Name: C-terminal LRRK2 bound to MLi-2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137 KDa |
-Macromolecule #1: Leucine-rich repeat serine/threonine-protein kinase 2
| Macromolecule | Name: Leucine-rich repeat serine/threonine-protein kinase 2 / type: protein_or_peptide / ID: 1 Details: Some loops are missing due to the lack of cryo-EM density Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 136.060656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RMKLMIVGNT GSGKTTLLQQ LMKTKKSDLG MQSATVGIDV KDWPIQIRDK RKRDLVLNVW DFAGREEFYS THPHFMTQRA LYLAVYDLS KGQAEVDAMK PWLFNIKARA SSSPVILVGT HLDVSDEKQR KACMSKITKE LLNKRGFPAI RDYHFVNATE E SDALAKLR ...String: RMKLMIVGNT GSGKTTLLQQ LMKTKKSDLG MQSATVGIDV KDWPIQIRDK RKRDLVLNVW DFAGREEFYS THPHFMTQRA LYLAVYDLS KGQAEVDAMK PWLFNIKARA SSSPVILVGT HLDVSDEKQR KACMSKITKE LLNKRGFPAI RDYHFVNATE E SDALAKLR KTIINESLNF KIRDQLVVGQ LIPDCYVELE KIILSERKNV PIEFPVIDRK RLLQLVRENQ LQLDENELPH AV HFLNESG VLLHFQDPAL QLSDLYFVEP KWLCKIMAQI LTVKVEGCPK HPKGIISRRD VEKFLSKKRK FPKNYMSQYF KLL EKFQIA LPIGEEYLLV PSSLSDHRPV IELPHCENSE IIIRLYEMPY FPMGFWSRLI NRLLEISPYM LSGRERALRP NRMY WRQGI YLNWSPEAYC LVGSEVLDNH PESFLKITVP SCRKGCILLG QVVDHIDSLM EEWFPGLLEI DICGEGETLL KKWAL YSFN DGEEHQKILL DDLMKKAEEG DLLVNPDQPR LTIPISQIAP DLILADLPRN IMLNNDELEF EQAPEFLLGD GSFGSV YRA AYEGEEVAVK IFNKHTSLRL LRQELVVLCH LHHPSLISLL AAGIRPRMLV MELASKGSLD RLLQQDKASL TRTLQHR IA LHVADGLRYL HSAMIIYRDL KPHNVLLFTL YPNAAIIAKI ADYGIAQYCC RMGIKTSEGT PGFRAPEVAR GNVIYNQQ A DVYSFGLLLY DILTTGGRIV EGLKFPNEFD ELEIQGKLPD PVKEYGCAPW PMVEKLIKQC LKENPQERPT SAQVFDILN SAELVCLTRR ILLPKNVIVE CMVATHHNSR NASIWLGCGH TDRGQLSFLD LNTEGYTSEE VADSRILCLA LVHLPVEKES WIVSGTQSG TLLVINTEDG KKRHTLEKMT DSVTCLYCNS FSKQSKQKNF LLVGTADGKL AIFEDKTVKL KGAAPLKILN I GNVSTPLM CLSESTNSTE RNVMWGGCGT KIFSFSNDFT IQKLIETRTS QLFSYAAFSD SNIITVVVDT ALYIAKQNSP VV EVWDKKT EKLCGLIDCV HFLREVMVKE NKESKHKMSY SGRVKTLCLQ KNTALWIGTG GGHILLLDLS TRRLIRVIYN FCN SVRVMM TAQLGSLKNV MLVLGYNRKN TEGTQKQKEI QSCLTVWDIN LPHEVQNLEK HIEVRKELAE KMRRTSVE UniProtKB: Leucine-rich repeat serine/threonine-protein kinase 2 |
-Macromolecule #2: (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}...
| Macromolecule | Name: (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine type: ligand / ID: 2 / Number of copies: 1 / Formula: A1N |
|---|---|
| Molecular weight | Theoretical: 379.456 Da |
| Chemical component information |  ChemComp-A1N: |
-Macromolecule #3: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.822 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||||||||
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Blot force = 3 Blot time = 4 seconds wait time = 20 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 11181 / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)