[English] 日本語
 Yorodumi
Yorodumi- EMDB-41795: Structure of C-terminal half of LRRK2 (I2020T mutant) bound to GZ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of C-terminal half of LRRK2 (I2020T mutant) bound to GZD-824, ROC-COR domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / GTPases / inhibitor / Parkinson / PROTEIN BINDING | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
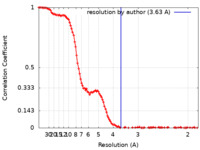
| Method | single particle reconstruction / cryo EM / Resolution: 3.63 Å | |||||||||
 Authors Authors | Villagran-Suarez A / Sanz-Murillo M / Alegrio-Louro J / Leschziner A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures. Authors: Marta Sanz Murillo / Amalia Villagran Suarez / Verena Dederer / Deep Chatterjee / Jaime Alegrio Louro / Stefan Knapp / Sebastian Mathea / Andres E Leschziner /   Abstract: Mutations in leucine-rich repeat kinase 2 (LRRK2) are a common cause of familial Parkinson's disease (PD) and a risk factor for the sporadic form. Increased kinase activity was shown in patients with ...Mutations in leucine-rich repeat kinase 2 (LRRK2) are a common cause of familial Parkinson's disease (PD) and a risk factor for the sporadic form. Increased kinase activity was shown in patients with both familial and sporadic PD, making LRRK2 kinase inhibitors a major focus of drug development efforts. Although much progress has been made in understanding the structural biology of LRRK2, there are no available structures of LRRK2 inhibitor complexes. To this end, we solved cryo-electron microscopy structures of LRRK2, wild-type and PD-linked mutants, bound to the LRRK2-specific type I inhibitor MLi-2 and the broad-spectrum type II inhibitor GZD-824. Our structures revealed an active-like LRRK2 kinase in the type I inhibitor complex, and an inactive DYG-out in the type II inhibitor complex. Our structural analysis also showed how inhibitor-induced conformational changes in LRRK2 are affected by its autoinhibitory N-terminal repeats. The structures provide a template for the rational development of LRRK2 kinase inhibitors covering both canonical inhibitor binding modes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41795.map.gz emd_41795.map.gz | 115.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41795-v30.xml emd-41795-v30.xml emd-41795.xml emd-41795.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41795_fsc.xml emd_41795_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_41795.png emd_41795.png | 30.7 KB | ||
| Filedesc metadata |  emd-41795.cif.gz emd-41795.cif.gz | 6.7 KB | ||
| Others |  emd_41795_half_map_1.map.gz emd_41795_half_map_1.map.gz emd_41795_half_map_2.map.gz emd_41795_half_map_2.map.gz | 113.6 MB 113.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41795 http://ftp.pdbj.org/pub/emdb/structures/EMD-41795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41795 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41795.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41795.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.935 Å | ||||||||||||||||||||||||||||||||||||
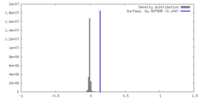
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41795_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
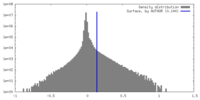
| Density Histograms |
-Half map: #1
| File | emd_41795_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
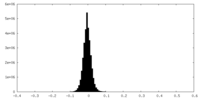
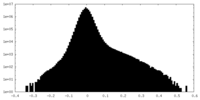
| Density Histograms |
- Sample components
Sample components
-Entire : C-terminal half of LRRK2 (I2020T mutant) bound to GZD-824 and DAR...
| Entire | Name: C-terminal half of LRRK2 (I2020T mutant) bound to GZD-824 and DARPin E11 |
|---|---|
| Components |
|
-Supramolecule #1: C-terminal half of LRRK2 (I2020T mutant) bound to GZD-824 and DAR...
| Supramolecule | Name: C-terminal half of LRRK2 (I2020T mutant) bound to GZD-824 and DARPin E11 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137 KDa |
-Macromolecule #1: C-terminal half of LRRK2 (I2020T mutant)
| Macromolecule | Name: C-terminal half of LRRK2 (I2020T mutant) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MASGSCQGCE EDEETLKKLI VRLNNVQEGK QIETLVQILE DLLVFTYSER ASKLFQGKNI HVPLLIVLDS YMRVASVQQV GWSLLCKLIE VCPGTMQSLM GPQDVGNDWE VLGVHQLILK MLTVHNASVN LSVIGLKTLD LLLTSGKITL LILDEESDIF MLIFDAMHSF ...String: MASGSCQGCE EDEETLKKLI VRLNNVQEGK QIETLVQILE DLLVFTYSER ASKLFQGKNI HVPLLIVLDS YMRVASVQQV GWSLLCKLIE VCPGTMQSLM GPQDVGNDWE VLGVHQLILK MLTVHNASVN LSVIGLKTLD LLLTSGKITL LILDEESDIF MLIFDAMHSF PANDEVQKLG CKALHVLFER VSEEQLTEFV ENKDYMILLS ALTNFKDEEE IVLHVLHCLH SLAIPCNNVE VLMSGNVRCY NIVVEAMKAF PMSERIQEVS CCLLHRLTLG NFFNILVLNE VHEFVVKAVQ QYPENAALQI SALSCLALLT ETIFLNQDLE EKNENQENDD EGEEDKLFWL EACYKALTWH RKNKHVQEAA CWALNNLLMY QNSLHEKIGD EDGHFPAHRE VMLSMLMHSS SKEVFQASAN ALSTLLEQNV NFRKILLSKG IHLNVLELMQ KHIHSPEVAE SGCKMLNHLF EGSNTSLDIM AAVVPKILTV MKRHETSLPV QLEALRAILH FIVPGMPEES REDTEFHHKL NMVKKQCFKN DIHKLVLAAL NRFIGNPGIQ KCGLKVISSI VHFPDALEML SLEGAMDSVL HTLQMYPDDQ EIQCLGLSLI GYLITKKNVF IGTGHLLAKI LVSSLYRFKD VAEIQTKGFQ TILAILKLSA SFSKLLVHHS FDLVIFHQMS SNIMEQKDQQ FLNLCCKCFA KVAMDDYLKN VMLERACDQN NSIMVECLLL LGADANQAKE GSSLICQVCE KESSPKLVEL LLNSGSREQD VRKALTISIG KGDSQIISLL LRRLALDVAN NSICLGGFCI GKVEPSWLGP LFPDKTSNLR KQTNIASTLA RMVIRYQMKS AVEEGTASGS DGNFSEDVLS KFDEWTFIPD SSMDSVFAQS DDLDSEGSEG SFLVKKKSNS ISVGEFYRDA VLQRCSPNLQ RHSNSLGPIF DHEDLLKRKR KILSSDDSLR SSKLQSHMRH SDSISSLASE REYITSLDLS ANELRDIDAL SQKCCISVHL EHLEKLELHQ NALTSFPQQL CETLKSLTHL DLHSNKFTSF PSYLLKMSCI ANLDVSRNDI GPSVVLDPTV KCPTLKQFNL SYNQLSFVPE NLTDVVEKLE QLILEGNKIS GICSPLRLKE LKILNLSKNH ISSLSENFLE ACPKVESFSA RMNFLAAMPF LPPSMTILKL SQNKFSCIPE AILNLPHLRS LDMSSNDIQY LPGPAHWKSL NLRELLFSHN QISILDLSEK AYLWSRVEKL HLSHNKLKEI PPEIGCLENL TSLDVSYNLE LRSFPNEMGK LSKIWDLPLD ELHLNFDFKH IGCKAKDIIR FLQQRLKKAV PYNRMKLMIV GNTGSGKTTL LQQLMKTKKS DLGMQSATVG IDVKDWPIQI RDKRKRDLVL NVWDFAGREE FYSTHPHFMT QRALYLAVYD LSKGQAEVDA MKPWLFNIKA RASSSPVILV GTHLDVSDEK QRKACMSKIT KELLNKRGFP AIRDYHFVNA TEESDALAKL RKTIINESLN FKIRDQLVVG QLIPDCYVEL EKIILSERKN VPIEFPVIDR KRLLQLVREN QLQLDENELP HAVHFLNESG VLLHFQDPAL QLSDLYFVEP KWLCKIMAQI LTVKVEGCPK HPKGIISRRD VEKFLSKKRK FPKNYMSQYF KLLEKFQIAL PIGEEYLLVP SSLSDHRPVI ELPHCENSEI IIRLYEMPYF PMGFWSRLIN RLLEISPYML SGRERALRPN RMYWRQGIYL NWSPEAYCLV GSEVLDNHPE SFLKITVPSC RKGCILLGQV VDHIDSLMEE WFPGLLEIDI CGEGETLLKK WALYSFNDGE EHQKILLDDL MKKAEEGDLL VNPDQPRLTI PISQIAPDLI LADLPRNIML NNDELEFEQA PEFLLGDGSF GSVYRAAYEG EEVAVKIFNK HTSLRLLRQE LVVLCHLHHP SLISLLAAGI RPRMLVMELA SKGSLDRLLQ QDKASLTRTL QHRIALHVAD GLRYLHSAMI IYRDLKPHNV LLFTLYPNAA IIAKIADYGI AQYCCRMGIK TSEGTPGFRA PEVARGNVIY NQQADVYSFG LLLYDILTTG GRIVEGLKFP NEFDELEIQG KLPDPVKEYG CAPWPMVEKL IKQCLKENPQ ERPTSAQVFD ILNSAELVCL TRRILLPKNV IVECMVATHH NSRNASIWLG CGHTDRGQLS FLDLNTEGYT SEEVADSRIL CLALVHLPVE KESWIVSGTQ SGTLLVINTE DGKKRHTLEK MTDSVTCLYC NSFSKQSKQK NFLLVGTADG KLAIFEDKTV KLKGAAPLKI LNIGNVSTPL MCLSESTNST ERNVMWGGCG TKIFSFSNDF TIQKLIETRT SQLFSYAAFS DSNIITVVVD TALYIAKQNS PVVEVWDKKT EKLCGLIDCV HFLREVMVKE NKESKHKMSY SGRVKTLCLQ KNTALWIGTG GGHILLLDLS TRRLIRVIYN FCNSVRVMMT AQLGSLKNVM LVLGYNRKNT EGTQKQKEIQ SCLTVWDINL PHEVQNLEKH IEVRKELAEK MRRTSVE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.625 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||||||||
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||||||||
| Details | 5uM LRRK2 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 2 / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)