[English] 日本語
 Yorodumi
Yorodumi- EMDB-33583: CryoEM structure of Klebsiella phage Kp7 tail complex applied wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Klebsiella phage Kp7 tail complex applied with C6 symmetry | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Podophage / short non-contractile tail / VIRUS | |||||||||
| Biological species |  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) | |||||||||
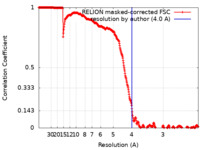
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Huang L / Xiang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure and assembly of the Klebsiella pneumoniae phage tail fibers Authors: Huang L / Xiang Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33583.map.gz emd_33583.map.gz | 106.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33583-v30.xml emd-33583-v30.xml emd-33583.xml emd-33583.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33583_fsc.xml emd_33583_fsc.xml | 35.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33583.png emd_33583.png | 28.2 KB | ||
| Filedesc metadata |  emd-33583.cif.gz emd-33583.cif.gz | 6.4 KB | ||
| Others |  emd_33583_half_map_1.map.gz emd_33583_half_map_1.map.gz emd_33583_half_map_2.map.gz emd_33583_half_map_2.map.gz | 3.1 GB 3.1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33583 http://ftp.pdbj.org/pub/emdb/structures/EMD-33583 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33583 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33583 | HTTPS FTP |
-Related structure data
| Related structure data |  7y22MC  7xy1C  7xycC  7y1cC  7y23C  7y3tC  7y5sC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33583.map.gz / Format: CCP4 / Size: 3.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33583.map.gz / Format: CCP4 / Size: 3.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.17 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33583_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33583_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Klebsiella phage Kp7
| Entire | Name:  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Klebsiella phage Kp7
| Supramolecule | Name: Klebsiella phage Kp7 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2936515 / Sci species name: Klebsiella phage Kp7 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Klebsiella pneumoniae ATCC 43816 (bacteria) Klebsiella pneumoniae ATCC 43816 (bacteria) |
| Virus shell | Shell ID: 1 / Diameter: 600.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: phage connector protein
| Macromolecule | Name: phage connector protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) |
| Molecular weight | Theoretical: 58.339309 KDa |
| Sequence | String: MDIRGGASYS GKKSKIPELW EKLSRKRNEF LDRAKHYARL TLPYLLNEPG NNESAQNGWQ GTGAQATNHL ANKLAQVLFP AQRSFFRVD LTVKGEKALL ERGYQKTKLA TVFAKIETQA MKALDARQFR PAVVEAFKHL LVAGNCMLYK PVKSANDPAG P ISCIPMHH ...String: MDIRGGASYS GKKSKIPELW EKLSRKRNEF LDRAKHYARL TLPYLLNEPG NNESAQNGWQ GTGAQATNHL ANKLAQVLFP AQRSFFRVD LTVKGEKALL ERGYQKTKLA TVFAKIETQA MKALDARQFR PAVVEAFKHL LVAGNCMLYK PVKSANDPAG P ISCIPMHH YAVQRDTNGQ LMDIILLQEK ALRTFEPALR AVIQAARKGK QLKDSDNVKL YTHACYEGDG FWKVQQSADD LP VGKSSRV KADKLPFMVL TWKRSYGEDW GRPLCEDYSG DLFVVQFLSE AVARGAALMA DIKYLIRPGA QTDVEHFVNS GTG EVITGV EEDIHIVQLG KYADLTPIDA VLEKYVRRIG VVFMMESLVR RDAERVTALE IQRDAMEVEQ SLGGAYSLFS VTMQ QPMAI WGLQENAGSF GSEFIDPVII TGIEALGRMA ELDKLAQFSN YMNLPTTWPE WAQDAIKPTE YMDWVRGQIS ADFPF LMSE EEAAAAAEQA QEQQADQTLN EGVAQAIPQV INQGLQEL |
-Macromolecule #2: phage tail tubular protein A
| Macromolecule | Name: phage tail tubular protein A / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) |
| Molecular weight | Theoretical: 27.228775 KDa |
| Sequence | String: MPIQSDTPSI MAESQFNSLS TKLDAVNLCM RAIGRSGVDN LTSGDLDAED ADTMIDIVSQ RLQYNDGKGW WFNREPRWSF APDSNGEVV LPNNTLQVLQ AYTLNTRKID ITIRAGRLYS TTLHSFDVSP LVGPDGFIWL DLMLMLPFEH MPLNVLQAIA Y QAAAEFIV ...String: MPIQSDTPSI MAESQFNSLS TKLDAVNLCM RAIGRSGVDN LTSGDLDAED ADTMIDIVSQ RLQYNDGKGW WFNREPRWSF APDSNGEVV LPNNTLQVLQ AYTLNTRKID ITIRAGRLYS TTLHSFDVSP LVGPDGFIWL DLMLMLPFEH MPLNVLQAIA Y QAAAEFIV SKDADQTKLQ MHMQMAANLH TGMQVEESKQ NRLNMLVHNP TQRNFGIMAG GPNNTAGFDH SPYDRYPVRP WR W |
-Macromolecule #3: phage tail tubular protein B
| Macromolecule | Name: phage tail tubular protein B / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) |
| Molecular weight | Theoretical: 87.904539 KDa |
| Sequence | String: MEVQGSLGRQ IQGISQQPAS VRLPGQCTDA INCSMDVVEG TKSRPGTVHI ARLGDLGLIQ DNTNIHHYRR GDDVEEYWMI TNPLGIPDI FDKQGRKCTV TETEGAASYF NSNNPRVDYK FFTVGDTTFV VNRTKIVRAR ADKTPAVGGT ALVFSAYGQY G TNYQIIIN ...String: MEVQGSLGRQ IQGISQQPAS VRLPGQCTDA INCSMDVVEG TKSRPGTVHI ARLGDLGLIQ DNTNIHHYRR GDDVEEYWMI TNPLGIPDI FDKQGRKCTV TETEGAASYF NSNNPRVDYK FFTVGDTTFV VNRTKIVRAR ADKTPAVGGT ALVFSAYGQY G TNYQIIIN GVKAAEYKTA SGGSASDVET IRTEVIAEQL YTNLLTWAGA SDYSISRMGT TIVISSLSGA SFTVDTEDGS KG KDLVAIQ YKVTSTDLLP SKAPVGYLVQ VWPTGSKPES RYWLKAEAAD GNLVTWQETL GADEVLGFDG STMPYIIERT NIV GGIAQF TIKQGYWDDR AVGDELTNPM PSFVDQSLSD IFMVQNRLCL AAGESCIMSR TSYFFQFFRQ TVLSAVDTDP IDVF ADASE VYALKHAKVL DGDTVLFSDN AQFILPGDKP LTKATALLRP TTTFEVDTNV APVVTGEAVM FATKDGAYSN IREFY TDSY SDTKKAQPVT SHVNKLIRGG IYHMASSTNF NRLFALSEDN RSRVFVYDWL WQGTDKVQSA WHKWEFYGAT IGGLYY SGE TLYLIIKRND GVFLEAMYMG DPLLSGSDQV RMDRTVTVSL TWDEATLSWK SSPLPWVPTQ VEMLEAVLTN GDPAYLG GA FLFEYDANTR ILSTKYGLGD TSQIWAAKVG QMYKVEFVPT DVIIRDSQDR VSYQDVPVIG LVHLNLDRYP DFTVEITN R KSGAVRVAKA SNRVGGARNN VVGYVKPTSG TFSFPLRALS TDVEYRIISI SPHTFQLRDI EWSGSYNPTR KRV |
-Macromolecule #4: tail adaptor protein
| Macromolecule | Name: tail adaptor protein / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) |
| Molecular weight | Theoretical: 36.062914 KDa |
| Sequence | String: MSYSYVERTG DGVATTFNFA FTGKGKGYLL ANQIYVERWD GASWQSATGW SLSGTNQITF LTPLANGQVI RIRRIAGKDY PFAQFEPGV MLDMASLNNT FIHLLEITQE LLDGFYPDGF YLKQDLNMGW NKIVNLMPGT DGGHAVNKTQ LDTLSSHVDD V DQKHTIWN ...String: MSYSYVERTG DGVATTFNFA FTGKGKGYLL ANQIYVERWD GASWQSATGW SLSGTNQITF LTPLANGQVI RIRRIAGKDY PFAQFEPGV MLDMASLNNT FIHLLEITQE LLDGFYPDGF YLKQDLNMGW NKIVNLMPGT DGGHAVNKTQ LDTLSSHVDD V DQKHTIWN DRQDQQIDGL LKAFDSNISY RTAPWTYEAA GGETMVFPPF YFASALVWRD GAYQDQQAGA FEIDNNVITL AD PPLRAGE RVSVLVGSYI TPADPGSWEW IHVAANGTTT SVDLGVSVSD IDDVTLDGLS QGRSNYTLTG TVLDFGEVIP ECT VGARVQ LA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 36000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 47 |
|---|---|
| Output model |  PDB-7y22: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)