[English] 日本語
 Yorodumi
Yorodumi- EMDB-33507: Cryo-EM structure of Klebsiella phage Kp9 type I tail fiber gp42 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Klebsiella phage Kp9 type I tail fiber gp42 in vitro | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Phage type I tail fiber / VIRAL PROTEIN | |||||||||
| Biological species |  Klebsiella phage Kp9 (virus) Klebsiella phage Kp9 (virus) | |||||||||
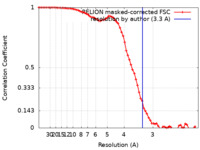
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Huang L / Xiang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure and assembly of the Klebsiella pneumoniae phage tail fibers Authors: Huang L / Xiang Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33507.map.gz emd_33507.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33507-v30.xml emd-33507-v30.xml emd-33507.xml emd-33507.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33507_fsc.xml emd_33507_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33507.png emd_33507.png | 15.4 KB | ||
| Filedesc metadata |  emd-33507.cif.gz emd-33507.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33507 http://ftp.pdbj.org/pub/emdb/structures/EMD-33507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33507 | HTTPS FTP |
-Related structure data
| Related structure data |  7xy1MC  7xycC  7y1cC  7y22C  7y23C  7y3tC  7y5sC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33507.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33507.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0742 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Klebsiella phage Kp9 type I tail fiber protein gp42
| Entire | Name: Klebsiella phage Kp9 type I tail fiber protein gp42 |
|---|---|
| Components |
|
-Supramolecule #1: Klebsiella phage Kp9 type I tail fiber protein gp42
| Supramolecule | Name: Klebsiella phage Kp9 type I tail fiber protein gp42 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Trimer of tail fiber proteins |
|---|---|
| Source (natural) | Organism:  Klebsiella phage Kp9 (virus) Klebsiella phage Kp9 (virus) |
-Macromolecule #1: Tail fiber protein
| Macromolecule | Name: Tail fiber protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella phage Kp9 (virus) Klebsiella phage Kp9 (virus) |
| Molecular weight | Theoretical: 84.120445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDQDIKTVIQ YPVGTTEFDI PFDYLSRKFV RVSLVSDDNR RLLSNITEYR YVSKTRVKLL VATTGFDRVE IRRFTSASER IVDFSDGSV LRANDLNVSQ LQSAHIAEEA RDAALLAMPE DDAGNLDARN RKIVRLAPGE AGTDAINKNQ LDTTLGEAGG I LSEVKDLQ ...String: MDQDIKTVIQ YPVGTTEFDI PFDYLSRKFV RVSLVSDDNR RLLSNITEYR YVSKTRVKLL VATTGFDRVE IRRFTSASER IVDFSDGSV LRANDLNVSQ LQSAHIAEEA RDAALLAMPE DDAGNLDARN RKIVRLAPGE AGTDAINKNQ LDTTLGEAGG I LSEVKDLQ KDMEDYLQNW GDDTTAIRGV LWVYNQGSAV GGETSFVITK EGPVLAVPYI EINGSRQYRG WHYEYDLGSK TI TLAKPLS AGDLVVCTTA ETTLPLADSL AGPTGASQIG TANGLNVQIA LDNLRSGVNV LDFMTFAERA AVLNYTGTND NSE AFRKAF ATGSRQIIVP PGRYHVKDVE IPSKVKLFGT YSYKPYNVTS DASFGTDGTI IRKVAGADNM FLWNTACAAE GVMF DGRDR TSPAIQSKSG GKISVGFFKC GFYRFDRVGN RRGAYIGCSF QFCNFNQNNI GIYNTVDGNH IGCTINANKS HGVML ETGA NSNTFTNCRN EWNEGDNWNF YGATSIQVIN ELCDRAFGYG FRISNSSVTL INVNIRRSAR TAASGAASAQ IYFESS TLK MIGVNSSVGG DDTGGSITEP SPDYFFRMAG TSEGRLEISD SRLTGYTVGL ISGTARPSVI RVINSPGWED TINEGVA RI SGGRPYIGTM PTATGPANVS PAVLGLSCGG VNTYDNDMFD IHLTIRNTNN GGHNGAILTV LLYREGGAAR ATIVRVDS R SNAVGEGDVN STSADPQQVY QVSVEVTSND ASTFNLLVST KSDNSASYRF RAKVKP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 124 |
|---|---|
| Output model |  PDB-7xy1: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)