[English] 日本語
 Yorodumi
Yorodumi- EMDB-33519: CryoEM structure of Klebsiella phage Kp7 type II tail fiber gp52 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Klebsiella phage Kp7 type II tail fiber gp52 in vitro | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Phage type II tail fiber / VIRAL PROTEIN | |||||||||
| Biological species |  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) | |||||||||
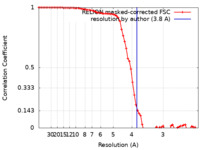
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Huang L / Xiang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure and assembly of the Klebsiella pneumoniae phage tail fibers Authors: Huang L / Xiang Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33519.map.gz emd_33519.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33519-v30.xml emd-33519-v30.xml emd-33519.xml emd-33519.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33519_fsc.xml emd_33519_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33519.png emd_33519.png | 37.2 KB | ||
| Filedesc metadata |  emd-33519.cif.gz emd-33519.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33519 http://ftp.pdbj.org/pub/emdb/structures/EMD-33519 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33519 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33519 | HTTPS FTP |
-Related structure data
| Related structure data |  7xycMC  7xy1C  7y1cC  7y22C  7y23C  7y3tC  7y5sC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33519.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33519.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.17 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Klebsiella phage Kp7 type II tail fiber
| Entire | Name: Klebsiella phage Kp7 type II tail fiber |
|---|---|
| Components |
|
-Supramolecule #1: Klebsiella phage Kp7 type II tail fiber
| Supramolecule | Name: Klebsiella phage Kp7 type II tail fiber / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Trimer of tail fiber proteins |
|---|---|
| Source (natural) | Organism:  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) |
-Macromolecule #1: phage tail fiber
| Macromolecule | Name: phage tail fiber / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella phage Kp7 (virus) Klebsiella phage Kp7 (virus) |
| Molecular weight | Theoretical: 61.21027 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALVKATFVK DVDGQPWRFS SVAKMKAFNY SCYLGSSVFL ESWHEGAGLG SGLFKVSKGT TEVGDDGSVI VAADGTRLIR VFDGPIFAD MWGALPSSTY NSLPAIKSAY LYASSKLQQL FLGGGSYKVT GSSGIDIDPS LAGISALSRA RVDTTEFTGD Y LFTITSSY ...String: MALVKATFVK DVDGQPWRFS SVAKMKAFNY SCYLGSSVFL ESWHEGAGLG SGLFKVSKGT TEVGDDGSVI VAADGTRLIR VFDGPIFAD MWGALPSSTY NSLPAIKSAY LYASSKLQQL FLGGGSYKVT GSSGIDIDPS LAGISALSRA RVDTTEFTGD Y LFTITSSY SYTPAPYYNN LSVALEGLYV FGNKTEGRSG LLTGRRTTDG VKSYNGQTEI RNCTFDKFDY NIRMGHNSWR IV FYKVNSL NALNANGILY VPSGLDDSGE ILTFYHCQFF DGAGSNIRIS CSSFAMTFVS CSFLNITFTI DAGSSVSVTA LGC NFENPG SQSTRRYIEI TAGHTNIFNV VGGSIVTNAN AGQTQALIHV SANNQINLSN LTIPYGAHYQ QEADSGYHAF CSGQ GYVST SNCSLQLLNG AGCCPIHPSL SVFTNWNLSY ANLNAWTVDK GSAPTSVAEY LSAQGPKGEG VLHVAPTTQG VNISQ VATV SKQAGSMSMS VMVNIISASS NAGQISLAYL DAFDNNLGGV SANLGTTTGW KVIGKNTLRG RLPVGTAKVR LNIQTV AGA DVQYTNILCN II |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 36000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 146 |
|---|---|
| Output model |  PDB-7xyc: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)