[English] 日本語
 Yorodumi
Yorodumi- EMDB-3121: cryoEM reconstruction of bnAb PGT128 in complex with BG505 SOSIP.... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3121 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM reconstruction of bnAb PGT128 in complex with BG505 SOSIP.664 Env trimer at 4.4 Angstrom resolution | |||||||||
 Map data Map data | Reconstruction of PGT128 Fab in complex with BG505 SOSIP.664 Env trimer at 4.4 Angstrom resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / Env / bnAb / antibody / PGT128 | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.36 Å | |||||||||
 Authors Authors | Lee JH / de Val N / Lyumkis D / Ward AB | |||||||||
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy. Authors: Jeong Hyun Lee / Natalia de Val / Dmitry Lyumkis / Andrew B Ward /  Abstract: Secretory and membrane proteins from mammalian cells undergo post-translational modifications, including N-linked glycosylation, which can result in a large number of possible glycoforms. This sample ...Secretory and membrane proteins from mammalian cells undergo post-translational modifications, including N-linked glycosylation, which can result in a large number of possible glycoforms. This sample heterogeneity can be problematic for structural studies, particularly X-ray crystallography. Thus, crystal structures of heavily glycosylated proteins such as the HIV-1 Env viral spike protein have been determined by removing the majority of glycans. This step is most frequently carried out using Endoglycosidase H (EndoH) and requires that all expressed glycans be in the high-mannose form, which is often not the native glycoform. With significantly improved technologies in single-particle cryoelectron microscopy, we demonstrate that it is now possible to refine and build natively glycosylated HIV-1 Env structures in solution to 4.36 Å resolution. At this resolution we can now analyze the complete epitope of a broadly neutralizing antibody (bnAb), PGT128, in the context of the trimer expressed with native glycans. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3121.map.gz emd_3121.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3121-v30.xml emd-3121-v30.xml emd-3121.xml emd-3121.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3121.png emd_3121.png | 346.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3121 http://ftp.pdbj.org/pub/emdb/structures/EMD-3121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3121 | HTTPS FTP |
-Validation report
| Summary document |  emd_3121_validation.pdf.gz emd_3121_validation.pdf.gz | 272.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3121_full_validation.pdf.gz emd_3121_full_validation.pdf.gz | 271.5 KB | Display | |
| Data in XML |  emd_3121_validation.xml.gz emd_3121_validation.xml.gz | 5.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3121 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3121 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3121 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3121 | HTTPS FTP |
-Related structure data
| Related structure data |  5acoMC  3120C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3121.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3121.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of PGT128 Fab in complex with BG505 SOSIP.664 Env trimer at 4.4 Angstrom resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
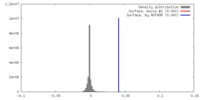
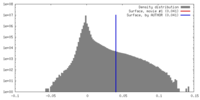
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PGT128 Fab bound to BG505 SOSIP.664 HIV-1 Env trimer
| Entire | Name: PGT128 Fab bound to BG505 SOSIP.664 HIV-1 Env trimer |
|---|---|
| Components |
|
-Supramolecule #1000: PGT128 Fab bound to BG505 SOSIP.664 HIV-1 Env trimer
| Supramolecule | Name: PGT128 Fab bound to BG505 SOSIP.664 HIV-1 Env trimer / type: sample / ID: 1000 Oligomeric state: Three monomers of PGT128 Fab bind one Env trimer Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: HIV-1 Envelope glycoprotein
| Macromolecule | Name: HIV-1 Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: Env Details: The Env sequence is from the clade A virus BG505, truncated at residue 664 of gp41, and contains stabilizing SOSIP mutations. Number of copies: 3 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: BG505 / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: BG505 / synonym: HIV-1 |
| Molecular weight | Theoretical: 420 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Macromolecule #2: Immunoglobulin G PGT128
| Macromolecule | Name: Immunoglobulin G PGT128 / type: protein_or_peptide / ID: 2 / Name.synonym: IgG PGT128 Details: The fragment antigen binding (Fab) of PGT128 was used to form the complex. Number of copies: 3 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 500 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris, 150 mM NaCl, 0.675 mM DDM |
| Grid | Details: 400 mesh C-Flat CF-2/2-4C, plasma treated for 5 seconds |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 160 K / Instrument: HOMEMADE PLUNGER Method: Grids were frozen using a manual plunger at 4 degrees. |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Alignment procedure | Legacy - Astigmatism: Objective astigmatism was corrected at 22,500x magnification |
| Date | Oct 7, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 2111 / Average electron dose: 33 e/Å2 Details: Each full dose image is an aligned stack of frames recorded each using a dose of ~10 e-/Angstrom^2/sec. |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 22500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #2
Electron microscopy #2
| Microscopy ID | 2 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Alignment procedure | Legacy - Astigmatism: Objective astigmatism was corrected at 22,500x magnification |
| Date | Nov 7, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 2111 / Average electron dose: 35 e/Å2 Details: Each full dose image is an aligned stack of frames recorded each using a dose of ~10 e-/Angstrom^2/sec. |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 22500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 4.36 Å / Resolution method: OTHER / Software - Name: Imagic, Relion Details: At the end of refinement which gave the 4.47 Angstrom resolution structure, a mask excluding the Fab constant domain (flexible region) was applied, and refined for an additional 3 iterations. Number images used: 92095 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: H / Chain - #1 - Chain ID: L |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-5aco: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)