+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24814 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | M. xanthus encapsulin shell protein EncA with T=3 symmetry | |||||||||
 Map data Map data | EncA with T=3 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nanocage / encapsulin / iron storage / bacterial nano-compartment / CYTOSOLIC PROTEIN / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Type 1 encapsulin shell protein / Encapsulating protein for peroxidase / : / encapsulin nanocompartment / iron ion transport / intracellular iron ion homeostasis / Type 1 encapsulin shell protein EncA Function and homology information Function and homology information | |||||||||
| Biological species |  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) | |||||||||
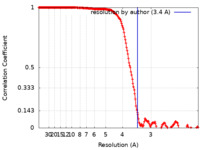
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Eren E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism. Authors: Elif Eren / Bing Wang / Dennis C Winkler / Norman R Watts / Alasdair C Steven / Paul T Wingfield /  Abstract: Encapsulins are bacterial organelle-like cages involved in various aspects of metabolism, especially protection from oxidative stress. They can serve as vehicles for a wide range of medical ...Encapsulins are bacterial organelle-like cages involved in various aspects of metabolism, especially protection from oxidative stress. They can serve as vehicles for a wide range of medical applications. Encapsulin shell proteins are structurally similar to HK97 bacteriophage capsid protein and their function depends on the encapsulated cargos. The Myxococcus xanthus encapsulin system comprises EncA and three cargos: EncB, EncC, and EncD. EncB and EncC are similar to bacterial ferritins that can oxidize Fe to less toxic Fe. We analyzed EncA, EncB, and EncC by cryo-EM and X-ray crystallography. Cryo-EM shows that EncA cages can have T = 3 and T = 1 symmetry and that EncA T = 1 has a unique protomer arrangement. Also, we define EncB and EncC binding sites on EncA. X-ray crystallography of EncB and EncC reveals conformational changes at the ferroxidase center and additional metal binding sites, suggesting a mechanism for Fe oxidation and storage within the encapsulin shell. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24814.map.gz emd_24814.map.gz | 52.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24814-v30.xml emd-24814-v30.xml emd-24814.xml emd-24814.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24814_fsc.xml emd_24814_fsc.xml | 19.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24814.png emd_24814.png | 250.7 KB | ||
| Filedesc metadata |  emd-24814.cif.gz emd-24814.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24814 http://ftp.pdbj.org/pub/emdb/structures/EMD-24814 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24814 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24814 | HTTPS FTP |
-Related structure data
| Related structure data |  7s20MC  7s21C  7s2tC  7s4qC  7s5cC  7s5kC  7s8tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24814.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24814.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EncA with T=3 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EncA with T=3 symmetry
| Entire | Name: EncA with T=3 symmetry |
|---|---|
| Components |
|
-Supramolecule #1: EncA with T=3 symmetry
| Supramolecule | Name: EncA with T=3 symmetry / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) |
-Macromolecule #1: EncA
| Macromolecule | Name: EncA / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) |
| Molecular weight | Theoretical: 33.505074 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHMPL EPHFMPDFLG HAENPLREEE WARLNETVIQ VARRSLVGRR ILDIYGPLGA GVQTVPYDEF QGVSPGAVDI VGEQETAMV FTDARKFKTI PIIYKDFLLH WRDIEAARTH NMPLDVSAAA GAAALCAQQE DELIFYGDAR LGYEGLMTAN G RLTVPLGD ...String: MHHHHHHMPL EPHFMPDFLG HAENPLREEE WARLNETVIQ VARRSLVGRR ILDIYGPLGA GVQTVPYDEF QGVSPGAVDI VGEQETAMV FTDARKFKTI PIIYKDFLLH WRDIEAARTH NMPLDVSAAA GAAALCAQQE DELIFYGDAR LGYEGLMTAN G RLTVPLGD WTSPGGGFQA IVEATRKLNE QGHFGPYAVV LSPRLYSQLH RIYEKTGVLE IETIRQLASD GVYQSNRLRG ES GVVVSTG RENMDLAVSM DMVAAYLGAS RMNHPFRVLE ALLLRIKHPD AICTLEGAGA TERR UniProtKB: Type 1 encapsulin shell protein EncA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.3 / Details: 20 mM HEPES, pH 7.3, 150 mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 12 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7s20: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)