+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5917 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Encapsulin protein (EncA) from Myxococcus xanthus | |||||||||
 Map data Map data | Reconstruction of the encapsulin protein (EncA) from Myxococcus xanthus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | encapsulin / Myxococcus xanthus / HK-97 fold / protein-bound organelle / iron storage | |||||||||
| Function / homology |  Function and homology information Function and homology informationencapsulin nanocompartment / iron ion transport / peptidase activity / intracellular iron ion homeostasis / defense response to bacterium Similarity search - Function | |||||||||
| Biological species |  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) | |||||||||
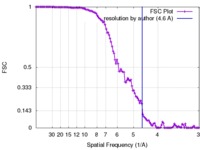
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | McHugh CA / Fontana J / Lam AS / Cheng N / Aksyuk AA / Steven AC / Hoiczyk E | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2014 Journal: EMBO J / Year: 2014Title: A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. Authors: Colleen A McHugh / Juan Fontana / Daniel Nemecek / Naiqian Cheng / Anastasia A Aksyuk / J Bernard Heymann / Dennis C Winkler / Alan S Lam / Joseph S Wall / Alasdair C Steven / Egbert Hoiczyk /  Abstract: Living cells compartmentalize materials and enzymatic reactions to increase metabolic efficiency. While eukaryotes use membrane-bound organelles, bacteria and archaea rely primarily on protein-bound ...Living cells compartmentalize materials and enzymatic reactions to increase metabolic efficiency. While eukaryotes use membrane-bound organelles, bacteria and archaea rely primarily on protein-bound nanocompartments. Encapsulins constitute a class of nanocompartments widespread in bacteria and archaea whose functions have hitherto been unclear. Here, we characterize the encapsulin nanocompartment from Myxococcus xanthus, which consists of a shell protein (EncA, 32.5 kDa) and three internal proteins (EncB, 17 kDa; EncC, 13 kDa; EncD, 11 kDa). Using cryo-electron microscopy, we determined that EncA self-assembles into an icosahedral shell 32 nm in diameter (26 nm internal diameter), built from 180 subunits with the fold first observed in bacteriophage HK97 capsid. The internal proteins, of which EncB and EncC have ferritin-like domains, attach to its inner surface. Native nanocompartments have dense iron-rich cores. Functionally, they resemble ferritins, cage-like iron storage proteins, but with a massively greater capacity (~30,000 iron atoms versus ~3,000 in ferritin). Physiological data reveal that few nanocompartments are assembled during vegetative growth, but they increase fivefold upon starvation, protecting cells from oxidative stress through iron sequestration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5917.map.gz emd_5917.map.gz | 317.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5917-v30.xml emd-5917-v30.xml emd-5917.xml emd-5917.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_5917_fsc.xml emd_5917_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_5917.png emd_5917.png | 291.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5917 http://ftp.pdbj.org/pub/emdb/structures/EMD-5917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5917 | HTTPS FTP |
-Related structure data
| Related structure data |  4pt2MC  5953C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5917.map.gz / Format: CCP4 / Size: 335 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5917.map.gz / Format: CCP4 / Size: 335 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the encapsulin protein (EncA) from Myxococcus xanthus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.102 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Encapsulin protein (EncA) from Myxococcus xanthus
| Entire | Name: Encapsulin protein (EncA) from Myxococcus xanthus |
|---|---|
| Components |
|
-Supramolecule #1000: Encapsulin protein (EncA) from Myxococcus xanthus
| Supramolecule | Name: Encapsulin protein (EncA) from Myxococcus xanthus / type: sample / ID: 1000 / Oligomeric state: Icosahedral / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 32 KDa |
-Macromolecule #1: Encapsulin
| Macromolecule | Name: Encapsulin / type: protein_or_peptide / ID: 1 / Name.synonym: EncA / Number of copies: 1 / Oligomeric state: Icosahedral / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Myxococcus xanthus (bacteria) / Strain: DK 1622 Myxococcus xanthus (bacteria) / Strain: DK 1622 |
| Molecular weight | Theoretical: 32 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Type 1 encapsulin shell protein EncA / GO: defense response to bacterium, peptidase activity / InterPro: Type 1 encapsulin shell protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 10 mM Tris, 10 mM MgCl2 |
| Grid | Details: 400 mesh carbon grid with thin carbon support |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 83 K / Instrument: LEICA KF80 |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Date | Dec 3, 2012 |
| Image recording | Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Number real images: 157 / Average electron dose: 15 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 57620 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.65 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)