+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21851 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | F. tularensis RNAPs70-(MglA-SspA)-iglA DNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase complex / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationsigma factor activity / glutathione metabolic process / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / transferase activity / protein dimerization activity / DNA-templated transcription ...sigma factor activity / glutathione metabolic process / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / transferase activity / protein dimerization activity / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Francisella tularensis subsp. holarctica LVS (bacteria) / Francisella tularensis subsp. holarctica LVS (bacteria) /  Francisella tularensis subsp. holarctica (strain LVS) (bacteria) Francisella tularensis subsp. holarctica (strain LVS) (bacteria) | |||||||||
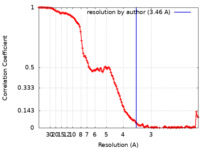
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Travis BA / Brennan RG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Structural Basis for Virulence Activation of Francisella tularensis. Authors: Brady A Travis / Kathryn M Ramsey / Samantha M Prezioso / Thomas Tallo / Jamie M Wandzilak / Allen Hsu / Mario Borgnia / Alberto Bartesaghi / Simon L Dove / Richard G Brennan / Maria A Schumacher /  Abstract: The bacterium Francisella tularensis (Ft) is one of the most infectious agents known. Ft virulence is controlled by a unique combination of transcription regulators: the MglA-SspA heterodimer, PigR, ...The bacterium Francisella tularensis (Ft) is one of the most infectious agents known. Ft virulence is controlled by a unique combination of transcription regulators: the MglA-SspA heterodimer, PigR, and the stress signal, ppGpp. MglA-SspA assembles with the σ-associated RNAP holoenzyme (RNAPσ), forming a virulence-specialized polymerase. These factors activate Francisella pathogenicity island (FPI) gene expression, which is required for virulence, but the mechanism is unknown. Here we report FtRNAPσ-promoter-DNA, FtRNAPσ-(MglA-SspA)-promoter DNA, and FtRNAPσ-(MglA-SspA)-ppGpp-PigR-promoter DNA cryo-EM structures. Structural and genetic analyses show MglA-SspA facilitates σ binding to DNA to regulate virulence and virulence-enhancing genes. Our Escherichia coli RNAPσhomodimeric EcSspA structure suggests this is a general SspA-transcription regulation mechanism. Strikingly, our FtRNAPσ-(MglA-SspA)-ppGpp-PigR-DNA structure reveals ppGpp binding to MglA-SspA tethers PigR to promoters. PigR in turn recruits FtRNAP αCTDs to DNA UP elements. Thus, these studies unveil a unique mechanism for Ft pathogenesis involving a virulence-specialized RNAP that employs two (MglA-SspA)-based strategies to activate virulence genes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21851.map.gz emd_21851.map.gz | 162.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21851-v30.xml emd-21851-v30.xml emd-21851.xml emd-21851.xml | 27.1 KB 27.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21851_fsc.xml emd_21851_fsc.xml | 16.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_21851.png emd_21851.png | 74.5 KB | ||
| Filedesc metadata |  emd-21851.cif.gz emd-21851.cif.gz | 8.7 KB | ||
| Others |  emd_21851_additional_1.map.gz emd_21851_additional_1.map.gz | 162.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21851 http://ftp.pdbj.org/pub/emdb/structures/EMD-21851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21851 | HTTPS FTP |
-Related structure data
| Related structure data |  6wmrMC  6wegC  6wmpC  6wmtC  6wmuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21851.map.gz / Format: CCP4 / Size: 175 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21851.map.gz / Format: CCP4 / Size: 175 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_21851_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Francisella RNAPs70-(MglA-SspA)-iglA complex
+Supramolecule #1: Francisella RNAPs70-(MglA-SspA)-iglA complex
+Macromolecule #1: DNA NT-strand
+Macromolecule #2: DNA T-strand
+Macromolecule #3: DNA NT-strand downstream
+Macromolecule #4: DNA T-strand downstream
+Macromolecule #5: Stringent starvation protein A
+Macromolecule #6: Macrophage growth locus, subunit A
+Macromolecule #7: RNA polymerase sigma factor RpoD
+Macromolecule #8: DNA-directed RNA polymerase subunit alpha 1
+Macromolecule #9: DNA-directed RNA polymerase subunit alpha 2
+Macromolecule #10: DNA-directed RNA polymerase subunit beta
+Macromolecule #11: DNA-directed RNA polymerase subunit beta'
+Macromolecule #12: DNA-directed RNA polymerase subunit omega
+Macromolecule #13: MAGNESIUM ION
+Macromolecule #14: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6wmr: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)