+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12309 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Trimeric efflux pump Klebsiella TolC in complex with KlebC | |||||||||
 Map data Map data | LocScale sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Efflux pump / trimer / complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationefflux pump complex / porin activity / efflux transmembrane transporter activity / cell outer membrane / response to antibiotic Similarity search - Function | |||||||||
| Biological species |  Klebsiella quasipneumoniae (bacteria) Klebsiella quasipneumoniae (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Webby MN / Housden NG | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Toxin import through the antibiotic efflux channel TolC. Authors: Nicholas G Housden / Melissa N Webby / Edward D Lowe / Tarick J El-Baba / Renata Kaminska / Christina Redfield / Carol V Robinson / Colin Kleanthous /  Abstract: Bacteria often secrete diffusible protein toxins (bacteriocins) to kill bystander cells during interbacterial competition. Here, we use biochemical, biophysical and structural analyses to show how a ...Bacteria often secrete diffusible protein toxins (bacteriocins) to kill bystander cells during interbacterial competition. Here, we use biochemical, biophysical and structural analyses to show how a bacteriocin exploits TolC, a major outer-membrane antibiotic efflux channel in Gram-negative bacteria, to transport itself across the outer membrane of target cells. Klebicin C (KlebC), a rRNase toxin produced by Klebsiella pneumoniae, binds TolC of a related species (K. quasipneumoniae) with high affinity through an N-terminal, elongated helical hairpin domain common amongst bacteriocins. The KlebC helical hairpin opens like a switchblade to bind TolC. A cryo-EM structure of this partially translocated state, at 3.1 Å resolution, reveals that KlebC associates along the length of the TolC channel. Thereafter, the unstructured N-terminus of KlebC protrudes beyond the TolC iris, presenting a TonB-box sequence to the periplasm. Association with proton-motive force-linked TonB in the inner membrane drives toxin import through the channel. Finally, we demonstrate that KlebC binding to TolC blocks drug efflux from bacteria. Our results indicate that TolC, in addition to its known role in antibiotic export, can function as a protein import channel for bacteriocins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12309.map.gz emd_12309.map.gz | 50 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12309-v30.xml emd-12309-v30.xml emd-12309.xml emd-12309.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12309_fsc.xml emd_12309_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_12309.png emd_12309.png | 39.7 KB | ||
| Filedesc metadata |  emd-12309.cif.gz emd-12309.cif.gz | 5.8 KB | ||
| Others |  emd_12309_additional_1.map.gz emd_12309_additional_1.map.gz emd_12309_additional_2.map.gz emd_12309_additional_2.map.gz emd_12309_additional_3.map.gz emd_12309_additional_3.map.gz | 57.2 MB 80.6 MB 9.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12309 http://ftp.pdbj.org/pub/emdb/structures/EMD-12309 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12309 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12309 | HTTPS FTP |
-Related structure data
| Related structure data |  7ng8MC  7ng9C  7nnaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12309.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12309.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocScale sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
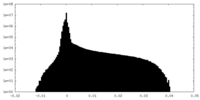
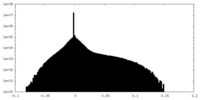
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened map from relion locres job. Requires Zflip
| File | emd_12309_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from relion locres job. Requires Zflip | ||||||||||||
| Projections & Slices |
| ||||||||||||
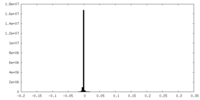
| Density Histograms |
-Additional map: Unsharpened map from relion refine3D job. Requires zflip
| File | emd_12309_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from relion refine3D job. Requires zflip | ||||||||||||
| Projections & Slices |
| ||||||||||||
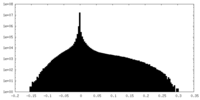
| Density Histograms |
-Additional map: Postprocess masked map from relion. Requires Zflip
| File | emd_12309_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess masked map from relion. Requires Zflip | ||||||||||||
| Projections & Slices |
| ||||||||||||
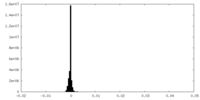
| Density Histograms |
- Sample components
Sample components
-Entire : Homotrimer of TolC from klebsiella in complex with KlebC
| Entire | Name: Homotrimer of TolC from klebsiella in complex with KlebC |
|---|---|
| Components |
|
-Supramolecule #1: Homotrimer of TolC from klebsiella in complex with KlebC
| Supramolecule | Name: Homotrimer of TolC from klebsiella in complex with KlebC type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Klebsiella quasipneumoniae (bacteria) Klebsiella quasipneumoniae (bacteria) |
-Macromolecule #1: Outer membrane channel protein
| Macromolecule | Name: Outer membrane channel protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella quasipneumoniae (bacteria) Klebsiella quasipneumoniae (bacteria) |
| Molecular weight | Theoretical: 54.058676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKLLPILIG LSLTGFSAMS QAENLLQVYQ QARISNPDLR KSAADRDAAF EKINEARSPL LPQLGLGADY TYTSGFRDYK DQNSNVTSG SLQLTQVLFD MSKWRALTLQ EKAAGIQDVT YQTDQQTLIL NTATAYFKVL AAIDTLSYTE AQKQAIYRQL D QTTQRFNV ...String: MKKLLPILIG LSLTGFSAMS QAENLLQVYQ QARISNPDLR KSAADRDAAF EKINEARSPL LPQLGLGADY TYTSGFRDYK DQNSNVTSG SLQLTQVLFD MSKWRALTLQ EKAAGIQDVT YQTDQQTLIL NTATAYFKVL AAIDTLSYTE AQKQAIYRQL D QTTQRFNV GLVAITDVQN ARSQYDAVLA NEVTARNDLD NAVEELRQVT GNYYPELASL NVDGFKTSKP QAVNALLKEA EN RNLSLLQ ARLNQDLARE QIRQAQDGHL PTLDLNASSG VSNNRYSGSK SISQDADIGQ NKIGLSFSLP LYQGGMVNSQ VKQ AQYNFV GASEQLESAH RSVVQTVRSS FNNVNASISS INAYKQAVVS AQSSLDAMEA GYSVGTRTIV DVLDATTTLY NAKQ QLSNA RYNYLINELN IKSALGTLNE QDLIALNNTL GKPISTSADS VAPENPQQDA TADGYGNTTA AMKPASARTT THSSG SNPF RQLEHHHHHH UniProtKB: Outer membrane protein TolC |
-Macromolecule #2: Klebicin C activity
| Macromolecule | Name: Klebicin C activity / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella quasipneumoniae (bacteria) Klebsiella quasipneumoniae (bacteria) |
| Molecular weight | Theoretical: 28.629162 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADNQPVPLT PAPPGMVSLG VNENGEEEMT VIGGDGSGTG FSGNEAPIIP GSGSLQADLG KKSLTRLQAE SSAAIHATAK WTTENLAKT QAAQAERAKA AMLSQQAAKA KQAKLTQHLK DVVDRALQNN KTRPTVIDLA HQNNQQMAAM AEFIGRQKAI E EARKKAER ...String: MADNQPVPLT PAPPGMVSLG VNENGEEEMT VIGGDGSGTG FSGNEAPIIP GSGSLQADLG KKSLTRLQAE SSAAIHATAK WTTENLAKT QAAQAERAKA AMLSQQAAKA KQAKLTQHLK DVVDRALQNN KTRPTVIDLA HQNNQQMAAM AEFIGRQKAI E EARKKAER EAKRAEEAYQ AALRAQEEEQ RKQAEIERKL QEARKQEAAA KAKAEADRIA AEKAEAEARA KAEAERRKAE EA RKALFAK AGIKDTPGCL EHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)