[English] 日本語
 Yorodumi
Yorodumi- EMDB-0488: Cryo-EM structure of the TRPM8 ion channel in complex with high o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0488 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | |||||||||
 Map data Map data | em-volume_P1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / TRP channel / TRPM channel / TRPM8 channel / cold sensing / lipid sensing / menthol / icilin / WS-12 / PI(4 / 5)P2 / cooling agent / MEMBRANE PROTEIN / calcium-permeable ion channel / TRANSPORT PROTEIN | |||||||||
| Biological species |  Ficedula albicollis (Collared flycatcher) Ficedula albicollis (Collared flycatcher) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Yin Y / Le SC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel. Authors: Ying Yin / Son C Le / Allen L Hsu / Mario J Borgnia / Huanghe Yang / Seok-Yong Lee /  Abstract: Transient receptor potential melastatin member 8 (TRPM8) is a calcium ion (Ca)-permeable cation channel that serves as the primary cold and menthol sensor in humans. Activation of TRPM8 by cooling ...Transient receptor potential melastatin member 8 (TRPM8) is a calcium ion (Ca)-permeable cation channel that serves as the primary cold and menthol sensor in humans. Activation of TRPM8 by cooling compounds relies on allosteric actions of agonist and membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP), but lack of structural information has thus far precluded a mechanistic understanding of ligand and lipid sensing by TRPM8. Using cryo-electron microscopy, we determined the structures of TRPM8 in complex with the synthetic cooling compound icilin, PIP, and Ca, as well as in complex with the menthol analog WS-12 and PIP Our structures reveal the binding sites for cooling agonists and PIP in TRPM8. Notably, PIP binds to TRPM8 in two different modes, which illustrate the mechanism of allosteric coupling between PIP and agonists. This study provides a platform for understanding the molecular mechanism of TRPM8 activation by cooling agents. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0488.map.gz emd_0488.map.gz | 58.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0488-v30.xml emd-0488-v30.xml emd-0488.xml emd-0488.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0488.png emd_0488.png | 265.2 KB | ||
| Filedesc metadata |  emd-0488.cif.gz emd-0488.cif.gz | 7.1 KB | ||
| Others |  emd_0488_half_map_1.map.gz emd_0488_half_map_1.map.gz emd_0488_half_map_2.map.gz emd_0488_half_map_2.map.gz | 45.6 MB 45.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0488 http://ftp.pdbj.org/pub/emdb/structures/EMD-0488 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0488 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0488 | HTTPS FTP |
-Related structure data
| Related structure data |  6nr3MC  0487C  0489C  6nr2C  6nr4C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0488.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0488.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-volume_P1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: em-half-volume P1
| File | emd_0488_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-half-volume_P1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: em-half-volume P2
| File | emd_0488_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-half-volume_P2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Transient receptor potential melastatin member 8
| Entire | Name: Transient receptor potential melastatin member 8 |
|---|---|
| Components |
|
-Supramolecule #1: Transient receptor potential melastatin member 8
| Supramolecule | Name: Transient receptor potential melastatin member 8 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Ficedula albicollis (Collared flycatcher) Ficedula albicollis (Collared flycatcher) |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 8
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 8 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ficedula albicollis (Collared flycatcher) Ficedula albicollis (Collared flycatcher) |
| Molecular weight | Theoretical: 130.011391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATLFQVSMG SMRHRRNGNF ESSRLLYSSM SRSIDVACSD ADLANFIQEN FKKRECVFFT KDTKSMGNLC KCGYPENQHI EGTQVNTTE KWNYKK(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) ...String: MATLFQVSMG SMRHRRNGNF ESSRLLYSSM SRSIDVACSD ADLANFIQEN FKKRECVFFT KDTKSMGNLC KCGYPENQHI EGTQVNTTE KWNYKK(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)K YIRLSCDTDS ETLYDLMTQH WHLKTPNLVI SVTG GAKNF ALKPRMRKIF SRLIYIAQSK GAWIFTGGTH YGLMKYIGEV VRDNTISRSS EENVVAIGIA AWGMISNRET LIRTA DSDG SYLAHYIMDD LKRDPLYCLD NNHTHLLLVD NGTHGHPTIE AKVRTQLEKY ISERVIPESN YGGKIPIVCF AQGGGK ETL KSINVAIKSK IPCVVVEGSG RIADVIASLV EAEGTLASSC VKESLLRFLP RTISRLSEEE TESWIKWIKE VLESPHL LT VIKIEEAGDE IVSNAISFAL YKAFSTNEHD RDNWNGQLKL LLEWNQLDLA SDEIFTNDRN WESADLQDVM FTALVKDR P KFVRLFLENG LNLRKFLTTE VLRELYTNNF SSLVFKNLQI AKNSYNDALL TFVWKMVEDF RRGAKRDDKN SKDEMEIEL SEECPITRHP LQALFIWSVL QNKKELSKVI WEQTRGCTLA ALGASKLLKS MAKVKNDINA AGESEELANE YETRAVELFT ECYSNDEDL AEQLLTYSCE AWGGSNCLEL AVEARDQQFI AQPGVQNFLS KQWYGEISRD TKNWKIILCL FFFPLIGCGF I SFRKKPVE KTKKLFLYYV SFFTSPFVVF SWNVIFYIAF LLLFAYVLLM DFQKEPTALE IILYVLVFIL LCDEVRQWYM NG SKYFSDL WNVMDTLGIF YFIAGIVFRL HSDESSWYSG RVIFCLDYIV FTLRLIHIFT VSRNLGPKII MLQRMMIDVF FFL FLFAVW MVAFGVARQG ILRKNEHRWE WIFRSVIYEP YLAMFGQYPD DIDGTTYNFD HCTFSGNESK PLCVELDANN QPRF PEWIT IPLVCIYMLS TNILLVNLLV AMFGYTVGSV QENNDQVWKF QRFFLVQEYC SRLTIPFPFV IFAYIFMVMR KCFKC CCKK ESKEPSVCCS RNEDNEILAW EAVMKENYLV KINTKASDSS EE(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)KI KSNSLEVLFQ GPDYKDDDDK AHHHHHHHHH H |
-Macromolecule #2: Icilin
| Macromolecule | Name: Icilin / type: ligand / ID: 2 / Number of copies: 4 / Formula: KX7 |
|---|---|
| Molecular weight | Theoretical: 311.292 Da |
| Chemical component information | 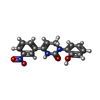 ChemComp-KX7: |
-Macromolecule #3: (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bi...
| Macromolecule | Name: (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate type: ligand / ID: 3 / Number of copies: 4 / Formula: KXP |
|---|---|
| Molecular weight | Theoretical: 1.047088 KDa |
| Chemical component information | 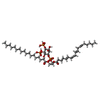 ChemComp-KXP: |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 26 sec. / Details: 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3705 / Average exposure time: 60.0 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.25 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)







































