+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-4058 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of the yeast spliceosome immediately after branching. Core without WD40 domain. | |||||||||
 マップデータ マップデータ | Spliceosome immediately after branching. Core without WD40. | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.1 Å | |||||||||
 データ登録者 データ登録者 | Galej WP / Wilkinson MF / Fica SM / Oubridge C / Newman AJ / Nagai K | |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2016 ジャーナル: Nature / 年: 2016タイトル: Cryo-EM structure of the spliceosome immediately after branching. 著者: Wojciech P Galej / Max E Wilkinson / Sebastian M Fica / Chris Oubridge / Andrew J Newman / Kiyoshi Nagai /  要旨: Precursor mRNA (pre-mRNA) splicing proceeds by two consecutive transesterification reactions via a lariat-intron intermediate. Here we present the 3.8 Å cryo-electron microscopy structure of the ...Precursor mRNA (pre-mRNA) splicing proceeds by two consecutive transesterification reactions via a lariat-intron intermediate. Here we present the 3.8 Å cryo-electron microscopy structure of the spliceosome immediately after lariat formation. The 5'-splice site is cleaved but remains close to the catalytic Mg site in the U2/U6 small nuclear RNA (snRNA) triplex, and the 5'-phosphate of the intron nucleotide G(+1) is linked to the branch adenosine 2'OH. The 5'-exon is held between the Prp8 amino-terminal and linker domains, and base-pairs with U5 snRNA loop 1. Non-Watson-Crick interactions between the branch helix and 5'-splice site dock the branch adenosine into the active site, while intron nucleotides +3 to +6 base-pair with the U6 snRNA ACAGAGA sequence. Isy1 and the step-one factors Yju2 and Cwc25 stabilize docking of the branch helix. The intron downstream of the branch site emerges between the Prp8 reverse transcriptase and linker domains and extends towards the Prp16 helicase, suggesting a plausible mechanism of remodelling before exon ligation. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_4058.map.gz emd_4058.map.gz | 243.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-4058-v30.xml emd-4058-v30.xml emd-4058.xml emd-4058.xml | 18.4 KB 18.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_4058_fsc.xml emd_4058_fsc.xml | 14.5 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_4058.png emd_4058.png | 45.4 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4058 http://ftp.pdbj.org/pub/emdb/structures/EMD-4058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4058 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_4058_validation.pdf.gz emd_4058_validation.pdf.gz | 309.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_4058_full_validation.pdf.gz emd_4058_full_validation.pdf.gz | 308.4 KB | 表示 | |
| XML形式データ |  emd_4058_validation.xml.gz emd_4058_validation.xml.gz | 14.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4058 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4058 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4058 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4058 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  4055C  4056C  4057C  4059C  5lj3C  5lj5C C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | |
| 電子顕微鏡画像生データ |  EMPIAR-10687 (タイトル: Yeast C, Ci, C*, and P complex spliceosomes / Data size: 8.9 TB EMPIAR-10687 (タイトル: Yeast C, Ci, C*, and P complex spliceosomes / Data size: 8.9 TBData #1: Unaligned movies of C-complex spliceosome with 3' splice site AG to AC mutation (Dataset 1) [micrographs - multiframe] Data #2: Unaligned movies of C and C*-complex spliceosomes with 3' splice site AG to AdG mutation (Dataset 2) [micrographs - multiframe] Data #3: Unaligned movies of C and C*-complex spliceosomes with 3' splice site AG to AdG mutation (Dataset 3) [micrographs - multiframe] Data #4: Aligned movies of C-complex spliceosomes with cold-sensitive prp16-302 mutation, purified with Cwc25 (Dataset 4) [micrographs - multiframe] Data #5: Unaligned movies of C-complex spliceosomes with cold-sensitive prp16-302 mutation, purified with Cwc25 and incubated with ATP and Mg (Dataset 5) [micrographs - multiframe] Data #6: Unaligned movies of C, C*, and P-complex spliceosomes with dominant-negative Prp22 mutation K512A, purified with Slu7 (Dataset 6) [micrographs - multiframe] Data #7: Unaligned movies of P-complex spliceosomes with dominant-negative Prp22 mutation K512A, treated with anti-3'exon RNaseH oligo, purified in presence of Mg (Dataset 9) [micrographs - single frame] Data #8: Selected C-complex particles after polishing [picked particles - single frame - processed] Data #9: Selected P-complex particles after polishing [picked particles - single frame - processed] Data #10: Various signal subtractions for C- and P-complex spliceosomes [picked particles - single frame - processed]) |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_4058.map.gz / 形式: CCP4 / 大きさ: 266.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_4058.map.gz / 形式: CCP4 / 大きさ: 266.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Spliceosome immediately after branching. Core without WD40. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.43 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
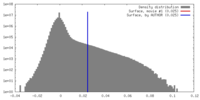
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Spliceosome immediately after branching
| 全体 | 名称: Spliceosome immediately after branching |
|---|---|
| 要素 |
|
-超分子 #1: Spliceosome immediately after branching
| 超分子 | 名称: Spliceosome immediately after branching / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#35 詳細: Splicing extract was prepared from Prp18-HA or Slu7-TAPS yeast strains. An in vitro transcribed yeast UBC4 pre-mRNA substrate (with 2 x MS2 bacteriophage coat protein-binding stem loops at ...詳細: Splicing extract was prepared from Prp18-HA or Slu7-TAPS yeast strains. An in vitro transcribed yeast UBC4 pre-mRNA substrate (with 2 x MS2 bacteriophage coat protein-binding stem loops at the 5' end and with the 3'-splice site sequence UAGAG mutated to UACAC) was pre-bound to an MS2-maltose binding protein fusion protein. This substrate-protein complex was added to the splicing extract. The splicing reaction proceeded through the first step but the second step was blocked by the 3' splice site mutation. Substrate-bound spliceosomes from the splicing extract were purified on amylose resin and eluted with maltose. Subsequently the spliceosomes were captured on anti-HA-agarose (for Prp18-HA-tagged) or streptactin resin (for Slu7-TAPS tagged) and eluted with HA peptide or desthiobiotin, respectively. Purified spliceosomes were then dialysed against 20 mM HEPES KOH pH 7.8, 75 mM KCl, 0.25 mM EDTA |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 2 MDa |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.8 構成要素:
| ||||||||||||
| グリッド | モデル: Quantifoil R2/2 / 材質: COPPER / メッシュ: 400 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS / 支持フィルム - Film thickness: 6.0 nm / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 雰囲気: AIR | ||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK III 詳細: 3 microlitres sample were applied to the grid, left for 30 seconds and then blotted for 2.5-3.0 seconds before plunging.. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: GIF Quantum |
| 撮影 | フィルム・検出器のモデル: GATAN K2 QUANTUM (4k x 4k) 検出モード: SUPER-RESOLUTION / デジタル化 - 画像ごとのフレーム数: 1-20 / 実像数: 2213 / 平均露光時間: 0.8 sec. / 平均電子線量: 2.0 e/Å2 詳細: Total dose: 40 electrons/Angstrom^2 over 16 seconds. 20 movie frames collected at 1.25 frames per second. |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 倍率(補正後): 35714 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 4.0 µm / 最小 デフォーカス(公称値): 0.5 µm / 倍率(公称値): 81000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 詳細 | Used secondary structure restraints generated in ProSMART and LibG. |
|---|---|
| 精密化 | 空間: RECIPROCAL / プロトコル: OTHER |
 ムービー
ムービー コントローラー
コントローラー










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)