[English] 日本語
 Yorodumi
Yorodumi- EMDB-30073: Cryo-EM structure of sulfur oxygenase reductase from Sulfurisphae... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30073 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | spherical homo 24-mer / OXIDOREDUCTASE | |||||||||
| Function / homology | sulfur oxygenase/reductase / sulfur oxygenase/reductase activity / Sulphur oxygenase reductase / Sulphur oxygenase reductase / Dimeric alpha-beta barrel / metal ion binding / Sulfur oxygenase/reductase Function and homology information Function and homology information | |||||||||
| Biological species |   Sulfurisphaera tokodaii (archaea) / Sulfurisphaera tokodaii (archaea) /   Sulfurisphaera tokodaii (strain DSM 16993 / JCM 10545 / NBRC 100140 / 7) (archaea) Sulfurisphaera tokodaii (strain DSM 16993 / JCM 10545 / NBRC 100140 / 7) (archaea) | |||||||||
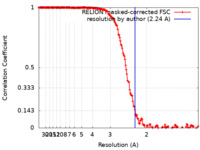
| Method | single particle reconstruction / cryo EM / Resolution: 2.24 Å | |||||||||
 Authors Authors | Sato Y / Adachi N | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol X / Year: 2020 Journal: J Struct Biol X / Year: 2020Title: Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase from . Authors: Yuta Sato / Takashi Yabuki / Naruhiko Adachi / Toshio Moriya / Takatoshi Arakawa / Masato Kawasaki / Chihaya Yamada / Toshiya Senda / Shinya Fushinobu / Takayoshi Wakagi /  Abstract: Sulfur oxygenase reductases (SORs) are present in thermophilic and mesophilic archaea and bacteria, and catalyze oxygen-dependent oxygenation and disproportionation of elemental sulfur. SOR has a ...Sulfur oxygenase reductases (SORs) are present in thermophilic and mesophilic archaea and bacteria, and catalyze oxygen-dependent oxygenation and disproportionation of elemental sulfur. SOR has a hollow, spherical homo-24-mer structure and reactions take place at active sites inside the chamber. The crystal structures of SORs from species have been reported. However, the states of the active site components (mononuclear iron and cysteines) and the entry and exit paths of the substrate and products are still in dispute. Here, we report the biochemical and structural characterizations of SORs from the thermoacidophilic archaeon (StSOR) and present high-resolution structures determined by X-ray crystallography and cryogenic electron microscopy (cryo-EM). The crystal structure of StSOR was determined at 1.73 Å resolution. At the catalytic center, iron is ligated to His86, His90, Glu114, and two water molecules. Three conserved cysteines in the cavity are located 9.5-13 Å from the iron and were observed as free thiol forms. A mutational analysis indicated that the iron and one of the cysteines (Cys31) were essential for both activities. The cryo-EM structure was determined at 2.24 Å resolution using an instrument operating at 200 kV. The two structures determined by different methodologies showed similar main chain traces, but the maps exhibited different features at catalytically important components. A possible role of StSOR in the sulfur metabolism of (an obligate aerobe) is discussed based on this study. Given the high resolution achieved in this study, StSOR was shown to be a good benchmark sample for cryo-EM. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30073.map.gz emd_30073.map.gz | 388 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30073-v30.xml emd-30073-v30.xml emd-30073.xml emd-30073.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30073_fsc.xml emd_30073_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_30073.png emd_30073.png | 321.7 KB | ||
| Filedesc metadata |  emd-30073.cif.gz emd-30073.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30073 http://ftp.pdbj.org/pub/emdb/structures/EMD-30073 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30073 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30073 | HTTPS FTP |
-Related structure data
| Related structure data |  6m3xMC  6m35C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10546 (Title: 2.05 angstrom resolution structure determination of sulfur oxygenase reductase using 200kV cryo-EM EMPIAR-10546 (Title: 2.05 angstrom resolution structure determination of sulfur oxygenase reductase using 200kV cryo-EMData size: 3.9 TB Data #1: 2.05 angstrom resolution structure determination of sulfur oxygenase reductase using 200kV cryo-EM [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30073.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30073.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.676 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : sulfur oxygenase reductase from Sulfurisphaera tokodaii
| Entire | Name: sulfur oxygenase reductase from Sulfurisphaera tokodaii |
|---|---|
| Components |
|
-Supramolecule #1: sulfur oxygenase reductase from Sulfurisphaera tokodaii
| Supramolecule | Name: sulfur oxygenase reductase from Sulfurisphaera tokodaii type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: spherical homo 24-mer |
|---|---|
| Source (natural) | Organism:   Sulfurisphaera tokodaii (archaea) / Strain: strain 7 Sulfurisphaera tokodaii (archaea) / Strain: strain 7 |
| Molecular weight | Theoretical: 857 KDa |
-Macromolecule #1: Sulfur oxygenase/reductase
| Macromolecule | Name: Sulfur oxygenase/reductase / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO / EC number: sulfur oxygenase/reductase |
|---|---|
| Source (natural) | Organism:   Sulfurisphaera tokodaii (strain DSM 16993 / JCM 10545 / NBRC 100140 / 7) (archaea) Sulfurisphaera tokodaii (strain DSM 16993 / JCM 10545 / NBRC 100140 / 7) (archaea)Strain: DSM 16993 / JCM 10545 / NBRC 100140 / 7 |
| Molecular weight | Theoretical: 35.763344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPKPYVAINM VEVRNDPKTL ELFGKVGPKV CMVTARHPGF VGFQNHVQIG VVPLGTRWGG AKMEMSQEMH SLMLMQYTFW KNWKDHEEM HKQNWANLFR LCLQCADQMI WGPYEPLYEI VYANMPLNTE MTDFTVMVGK KFAAGEAVSI PPISQPYGKR V VAFGEHIV ...String: MPKPYVAINM VEVRNDPKTL ELFGKVGPKV CMVTARHPGF VGFQNHVQIG VVPLGTRWGG AKMEMSQEMH SLMLMQYTFW KNWKDHEEM HKQNWANLFR LCLQCADQMI WGPYEPLYEI VYANMPLNTE MTDFTVMVGK KFAAGEAVSI PPISQPYGKR V VAFGEHIV KEGLENQFEE YAIKTLEAFR SAPGFLGGMI LKEIGVSPLG SLQLNAKGFH QILETANGMD VPEPVTIYEA PE FRNRPQR YIVHTEWSDT NALMFGLGRV LIYPEVRQIH DKVLDTLVYG PYIRVLNPMM EGTYWREYLN EYHL UniProtKB: Sulfur oxygenase/reductase |
-Macromolecule #2: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 2 / Number of copies: 24 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 2243 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: The grid was washed by acetone prior to use. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time was 5 seconds (blot force 20). | |||||||||
| Details | This sample was mono-disperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2558 / Average exposure time: 50.89 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 150000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 20.65 / Target criteria: CCmask |
|---|---|
| Output model |  PDB-6m3x: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)