[English] 日本語
 Yorodumi
Yorodumi- EMDB-25672: Structure of the C-terminal half of Leucine Rich Repeat Kinase 1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the C-terminal half of Leucine Rich Repeat Kinase 1 (LRRK1) | |||||||||
 Map data Map data | Catalytic C-terminus of Leucine Rich Repeat Kinase 1 (LRRK1) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | kinase / gtpase / CYTOSOLIC PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
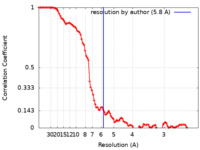
| Method | single particle reconstruction / cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | Matyszewski M / Snead DM / Leschziner AE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structural basis for Parkinson's disease-linked LRRK2's binding to microtubules. Authors: David M Snead / Mariusz Matyszewski / Andrea M Dickey / Yu Xuan Lin / Andres E Leschziner / Samara L Reck-Peterson /  Abstract: Leucine-rich repeat kinase 2 (LRRK2) is one of the most commonly mutated genes in familial Parkinson's disease (PD). Under some circumstances, LRRK2 co-localizes with microtubules in cells, an ...Leucine-rich repeat kinase 2 (LRRK2) is one of the most commonly mutated genes in familial Parkinson's disease (PD). Under some circumstances, LRRK2 co-localizes with microtubules in cells, an association enhanced by PD mutations. We report a cryo-EM structure of the catalytic half of LRRK2, containing its kinase, in a closed conformation, and GTPase domains, bound to microtubules. We also report a structure of the catalytic half of LRRK1, which is closely related to LRRK2 but is not linked to PD. Although LRRK1's structure is similar to that of LRRK2, we find that LRRK1 does not interact with microtubules. Guided by these structures, we identify amino acids in LRRK2's GTPase that mediate microtubule binding; mutating them disrupts microtubule binding in vitro and in cells, without affecting LRRK2's kinase activity. Our results have implications for the design of therapeutic LRRK2 kinase inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25672.map.gz emd_25672.map.gz | 85.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25672-v30.xml emd-25672-v30.xml emd-25672.xml emd-25672.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25672_fsc.xml emd_25672_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_25672.png emd_25672.png | 43.2 KB | ||
| Masks |  emd_25672_msk_1.map emd_25672_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25672.cif.gz emd-25672.cif.gz | 6.3 KB | ||
| Others |  emd_25672_additional_1.map.gz emd_25672_additional_1.map.gz emd_25672_half_map_1.map.gz emd_25672_half_map_1.map.gz emd_25672_half_map_2.map.gz emd_25672_half_map_2.map.gz | 45.3 MB 84.5 MB 84.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25672 http://ftp.pdbj.org/pub/emdb/structures/EMD-25672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25672 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25672.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25672.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Catalytic C-terminus of Leucine Rich Repeat Kinase 1 (LRRK1) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25672_msk_1.map emd_25672_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Non-sharpened map of LRRK1
| File | emd_25672_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened map of LRRK1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_25672_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_25672_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Catalytic C-terminus of Leucine Rich Repeat Kinase 1
| Entire | Name: Catalytic C-terminus of Leucine Rich Repeat Kinase 1 |
|---|---|
| Components |
|
-Supramolecule #1: Catalytic C-terminus of Leucine Rich Repeat Kinase 1
| Supramolecule | Name: Catalytic C-terminus of Leucine Rich Repeat Kinase 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 155 KDa |
-Macromolecule #1: C-terminal half of Leucine Rich Repeat Kinase 1
| Macromolecule | Name: C-terminal half of Leucine Rich Repeat Kinase 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: RKAEKCKLMK MIIVGPPRQG KSTLLEILQT GRAPQVVHGE ATIRTTKWEL QRPAGSRAKV ESVEFNVWD IGGPASMATV NQCFFTDKAL YVVVWNLALG EEAVANLQFW LLNIEAKAPN A VVLVVGTH LDLIEAKFRV ERIATLRAYV LALCRSPSGS RATGFPDITF ...String: RKAEKCKLMK MIIVGPPRQG KSTLLEILQT GRAPQVVHGE ATIRTTKWEL QRPAGSRAKV ESVEFNVWD IGGPASMATV NQCFFTDKAL YVVVWNLALG EEAVANLQFW LLNIEAKAPN A VVLVVGTH LDLIEAKFRV ERIATLRAYV LALCRSPSGS RATGFPDITF KHLHEISCKS LE GQEGLRQ LIFHVTCSMK DVGSTIGCQR LAGRLIPRSY LSLQEAVLAE QQRRSRDDDV QYL TDRQLE QLVEQTPDND IKDYEDLQSA ISFLIETGTL LHFPDTSHGL RNLYFLDPIW LSEC LQRIF NIKGSRSVAK NGVIRAEDLR MLLVGTGFTQ QTEEQYFQFL AKFEIALPVA NDSYL LPHL LPSKPGLDTH GMRHPTANTI QRVFKMSFVP VGFWQRFIAR MLISLAEMDL QLFENK KNT KSRNRKVTIY SFTGNQRNRC STFRVKRNQT IYWQEGLLVT FDGGYLSVES SDVNWKK KK SGGMKIVCQS EVRDFSAMAF ITDHVNSLID QWFPALTATE SDGTPLMEQY VPCPVCET A WAQHTDPSEK SEDVQYFDME DCVLTAIERD FISCPRHPDL PVPLQELVPE LFMTDFPAR LFLENSKLEH SEDEGSVLGQ GGSGTVIYRA RYQGQPVAVK RFHIKKFKNF ANVPADTMLR HLRATDAMK NFSEFRQEAS MLHALQHPCI VALIGISIHP LCFALELAPL SSLNTVLSEN A RDSSFIPL GHMLTQKIAY QIASGLAYLH KKNIIFCDLK SDNILVWSLD VKEHINIKLS DY GISRQSF HEGALGVEGT PGYQAPEIRP RIVYDEKVDM FSYGMVLYEL LSGQRPALGH HQL QIAKKL SKGIRPVLGQ PEEVQFRRLQ ALMMECWDTK PEKRPLALSV VSQMKDPTFA TFMY ELCCG KQTAFFSSQG QEYTVVFWDG KEESRNYTVV NTEKGLMEVQ RMCCPGMKVS CQLQV QRSL WTATEDQKIY IYTLKGMCPL NTPQQALDTP AVVTCFLAVP VIKKNSYLVL AGLADG LVA VFPVVRGTPK DSCSYLCSHT ANRSKFSIAD EDARQNPYPV KAMEVVNSGS EVWYSNG PG LLVIDCASLE ICRRLEPYMA PSMVTSVVCS SEGRGEEVVW CLDDKANSLV MYHSTTYQ L CARYFCGVPS PLRDMFPVRP LDTEPPAASH TANPKVPEGD SIADVSIMYS EELGTQILI HQESLTDYCS MSSYSSSPPR QAARSPSSLP SSPASSSSVP FSTDCEDSDM LHTPGAASDR SEHDLTPMD GETFSQHLQA VKILAVRDLI WVPRRGGDVI VIGLEKDSGA QRGRVIAVLK A RELTPHGV LVDAAVVAKD TVVCTFENEN TEWCLAVWRG WGAREFDIFY QSYEELGRLE AC TRKRR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | Concentrations varied from 2 to 6 uM depending on the grid. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Details | 1 out of the 4 datasets collected was tilted by 20 degrees |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 4 / Average electron dose: 55.0 e/Å2 Details: 1 dataset was tilted by 20 degrees. 250 ms frames used |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 36000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)