[English] 日本語
 Yorodumi
Yorodumi- EMDB-24400: cryo-EM structure of the rigor state Jordan myosin-15-F-actin complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24400 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of the rigor state Jordan myosin-15-F-actin complex | ||||||||||||
 Map data Map data | the rigor state Jordan myosin-15-F-actin complex | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | myosin motor proteins / actin cytoskeleton / stereocilia / deafness / MOTOR PROTEIN / MOTOR PROTEIN-ATP Binding Protein complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationStriated Muscle Contraction / stereocilium bundle / stereocilium / myosin complex / inner ear morphogenesis / cytoskeletal motor activity / striated muscle thin filament / skeletal muscle thin filament assembly / response to light stimulus / skeletal muscle fiber development ...Striated Muscle Contraction / stereocilium bundle / stereocilium / myosin complex / inner ear morphogenesis / cytoskeletal motor activity / striated muscle thin filament / skeletal muscle thin filament assembly / response to light stimulus / skeletal muscle fiber development / stress fiber / actin filament / locomotory behavior / sensory perception of sound / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / actin cytoskeleton / actin binding / hydrolase activity / ATP binding / cytosol Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
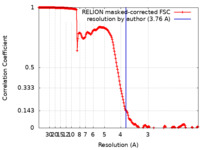
| Method | helical reconstruction / cryo EM / Resolution: 3.76 Å | ||||||||||||
 Authors Authors | Gong R / Reynolds MJ | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia. Authors: Rui Gong / Fangfang Jiang / Zane G Moreland / Matthew J Reynolds / Santiago Espinosa de Los Reyes / Pinar Gurel / Arik Shams / James B Heidings / Michael R Bowl / Jonathan E Bird / Gregory M Alushin /   Abstract: The motor protein myosin-15 is necessary for the development and maintenance of mechanosensory stereocilia, and mutations in myosin-15 cause hereditary deafness. In addition to transporting actin ...The motor protein myosin-15 is necessary for the development and maintenance of mechanosensory stereocilia, and mutations in myosin-15 cause hereditary deafness. In addition to transporting actin regulatory machinery to stereocilia tips, myosin-15 directly nucleates actin filament ("F-actin") assembly, which is disrupted by a progressive hearing loss mutation (p.D1647G, ""). Here, we present cryo-electron microscopy structures of myosin-15 bound to F-actin, providing a framework for interpreting the impacts of deafness mutations on motor activity and actin nucleation. Rigor myosin-15 evokes conformational changes in F-actin yet maintains flexibility in actin's D-loop, which mediates inter-subunit contacts, while the mutant locks the D-loop in a single conformation. Adenosine diphosphate-bound myosin-15 also locks the D-loop, which correspondingly blunts actin-polymerization stimulation. We propose myosin-15 enhances polymerization by bridging actin protomers, regulating nucleation efficiency by modulating actin's structural plasticity in a myosin nucleotide state-dependent manner. This tunable regulation of actin polymerization could be harnessed to precisely control stereocilium height. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia Authors: Gong R / Jiang F / Moreland ZG / Reynolds MJ / Gurel PS / Shams A / Bowl MR / Bird JE / Alushin GM | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24400.map.gz emd_24400.map.gz | 285.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24400-v30.xml emd-24400-v30.xml emd-24400.xml emd-24400.xml | 28.9 KB 28.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24400_fsc.xml emd_24400_fsc.xml | 18.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_24400.png emd_24400.png | 128.7 KB | ||
| Masks |  emd_24400_msk_1.map emd_24400_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24400.cif.gz emd-24400.cif.gz | 8.3 KB | ||
| Others |  emd_24400_half_map_1.map.gz emd_24400_half_map_1.map.gz emd_24400_half_map_2.map.gz emd_24400_half_map_2.map.gz | 410.5 MB 410.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24400 http://ftp.pdbj.org/pub/emdb/structures/EMD-24400 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24400 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24400 | HTTPS FTP |
-Related structure data
| Related structure data |  7rb9MC  7r8vC  7r91C  7rb8C  7udtC  7uduC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24400.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24400.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | the rigor state Jordan myosin-15-F-actin complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
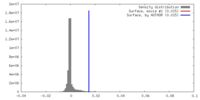
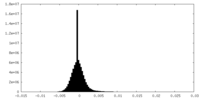
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24400_msk_1.map emd_24400_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
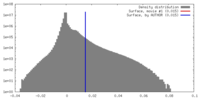
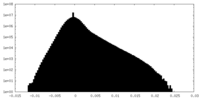
| Density Histograms |
-Half map: half1
| File | emd_24400_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
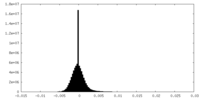
| ||||||||||||
| Density Histograms |
-Half map: half2
| File | emd_24400_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
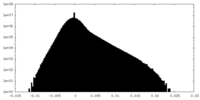
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : the rigor state Jordan myosin-15-F-actin complex
| Entire | Name: the rigor state Jordan myosin-15-F-actin complex |
|---|---|
| Components |
|
-Supramolecule #1: the rigor state Jordan myosin-15-F-actin complex
| Supramolecule | Name: the rigor state Jordan myosin-15-F-actin complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 5.4 kDa/nm |
-Supramolecule #2: Actin
| Supramolecule | Name: Actin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Isoform 3 of Unconventional myosin-XV
| Supramolecule | Name: Isoform 3 of Unconventional myosin-XV / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha skeletal muscle, intermediate form
| Macromolecule | Name: Actin, alpha skeletal muscle, intermediate form / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.63143 KDa |
| Sequence | String: DETTALVCDN GSGLVKAGFA GDDAPRAVFP SIVGRPRHQG VMVGMGQKDS YVGDEAQSKR GILTLKYPIE (HIC)GIITN WDD MEKIWHHTFY NELRVAPEEH PTLLTEAPLN PKANREKMTQ IMFETFNVPA MYVAIQAVLS LYASGRTTGI VLDSGDG VT HNVPIYEGYA ...String: DETTALVCDN GSGLVKAGFA GDDAPRAVFP SIVGRPRHQG VMVGMGQKDS YVGDEAQSKR GILTLKYPIE (HIC)GIITN WDD MEKIWHHTFY NELRVAPEEH PTLLTEAPLN PKANREKMTQ IMFETFNVPA MYVAIQAVLS LYASGRTTGI VLDSGDG VT HNVPIYEGYA LPHAIMRLDL AGRDLTDYLM KILTERGYSF VTTAEREIVR DIKEKLCYVA LDFENEMATA ASSSSLEK S YELPDGQVIT IGNERFRCPE TLFQPSFIGM ESAGIHETTY NSIMKCDIDI RKDLYANNVM SGGTTMYPGI ADRMQKEIT ALAPSTMKIK IIAPPERKYS VWIGGSILAS LSTFQQMWIT KQEYDEAGPS IVHRKCF UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #2: Isoform 3 of Unconventional myosin-XV
| Macromolecule | Name: Isoform 3 of Unconventional myosin-XV / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.124672 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) |
| Sequence | String: EDGVEDMTQL EDLQETTVLA NLKTRFERNL IYTYIGSILV SVNPYRMFAI YGPEQVQQYS GRALGENPPH LFAIANLAFA KMLDAKQNQ CVIISGESGS GKTEATKLIL RCLAAMNQRR DVMQQIKILE ATPLLEAFGN AKTVRNDNSS RFGKFVEIFL E GGVICGAI ...String: EDGVEDMTQL EDLQETTVLA NLKTRFERNL IYTYIGSILV SVNPYRMFAI YGPEQVQQYS GRALGENPPH LFAIANLAFA KMLDAKQNQ CVIISGESGS GKTEATKLIL RCLAAMNQRR DVMQQIKILE ATPLLEAFGN AKTVRNDNSS RFGKFVEIFL E GGVICGAI TSQYLLEKSR IVFQAKNERN YHIFYELLAG LPAQLRQAFS LQEAETYYYL NQGGNCEIAG KSDADDFRRL LA AMEVLGF TSEDQDSIFR ILASILHLGN VYFEKHETDA QEVASVVSAR EIQAVAELLQ VSPEGLQKAI TFKVTETIRE KIF TPLTVE SAVDARDAIA KVLYALLFGW LITRVNALVS PKQDTLSIAI LDIYGFEDLS FNSFEQLCIN YANENLQYLF NKIV FQEEQ EEYIREQMDW REIAFADNQP CINLISLKPY GILRILDGQC CFPQATDHTF LQKCHYHHGA NPLYSKPKMP LPEFT IKHY AGKVTYQVHK FLDKNHDQVR QDVLDLFVHS RTRVVAHLFS SHAAQTAPPR LGKSSSITRL YKAHTVAAKF QQSLLD LVE KMERCNPLFV RCLKPNHKKE PGLFEPDVMM AQLRYSGVLE TVRIRKEGFP VRLPFQVFID RYRCLVALKL NVPADGD MC VSLLSRLCTV TPDMYRVGIS KLFLKEHLHQ LLESMRERVQ NRA UniProtKB: Unconventional myosin-XV |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Details: 10 mM imidazole pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, 0.01% NaN3 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)