+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21847 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human V-ATPase in state 1 with SidK and ADP | |||||||||
 Map data Map data | Human V-ATPase in state 1 (composite map) | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationproton-transporting two-sector ATPase complex / Blockage of phagosome acidification / Ion channel transport / Regulation of MITF-M-dependent genes involved in lysosome biogenesis and autophagy / intracellular pH reduction / eye pigmentation / central nervous system maturation / Nef Mediated CD8 Down-regulation / ATPase-coupled ion transmembrane transporter activity / transporter activator activity ...proton-transporting two-sector ATPase complex / Blockage of phagosome acidification / Ion channel transport / Regulation of MITF-M-dependent genes involved in lysosome biogenesis and autophagy / intracellular pH reduction / eye pigmentation / central nervous system maturation / Nef Mediated CD8 Down-regulation / ATPase-coupled ion transmembrane transporter activity / transporter activator activity / plasma membrane proton-transporting V-type ATPase complex / rostrocaudal neural tube patterning / positive regulation of transforming growth factor beta1 production / cellular response to increased oxygen levels / Golgi lumen acidification / proton-transporting V-type ATPase, V0 domain / synaptic vesicle lumen acidification / Transferrin endocytosis and recycling / extrinsic component of synaptic vesicle membrane / lysosomal lumen acidification / clathrin-coated vesicle membrane / endosome to plasma membrane protein transport / vacuolar transport / endosomal lumen acidification / vacuolar proton-transporting V-type ATPase, V0 domain / vacuolar proton-transporting V-type ATPase, V1 domain / XBP1(S) activates chaperone genes / vacuolar proton-transporting V-type ATPase complex / Amino acids regulate mTORC1 / proton-transporting V-type ATPase complex / head morphogenesis / vacuolar acidification / ROS and RNS production in phagocytes / osteoclast development / protein localization to cilium / Nef Mediated CD4 Down-regulation / dendritic spine membrane / regulation of cellular pH / azurophil granule membrane / regulation of MAPK cascade / microvillus / proton transmembrane transporter activity / tertiary granule membrane / ATPase activator activity / autophagosome membrane / positive regulation of Wnt signaling pathway / ficolin-1-rich granule membrane / cilium assembly / RHOA GTPase cycle / regulation of macroautophagy / angiotensin maturation / Metabolism of Angiotensinogen to Angiotensins / specific granule membrane / enzyme regulator activity / H+-transporting two-sector ATPase / ATP metabolic process / axon terminus / Insulin receptor recycling / ruffle / endoplasmic reticulum-Golgi intermediate compartment membrane / proton-transporting ATPase activity, rotational mechanism / RNA endonuclease activity / proton-transporting ATP synthase activity, rotational mechanism / receptor-mediated endocytosis / proton transmembrane transport / secretory granule membrane / secretory granule / transmembrane transport / cilium / small GTPase binding / synaptic vesicle membrane / endocytosis / phagocytic vesicle membrane / positive regulation of canonical Wnt signaling pathway / melanosome / apical part of cell / signaling receptor activity / ATPase binding / postsynaptic membrane / intracellular iron ion homeostasis / Hydrolases; Acting on ester bonds / receptor-mediated endocytosis of virus by host cell / lysosome / early endosome / endosome membrane / endosome / nuclear speck / apical plasma membrane / lysosomal membrane / Golgi membrane / axon / external side of plasma membrane / intracellular membrane-bounded organelle / focal adhesion / centrosome / ubiquitin protein ligase binding / Neutrophil degranulation / endoplasmic reticulum membrane / protein-containing complex binding / perinuclear region of cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Human (human) / Human (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Wang L / Wu H / Fu TM | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly. Authors: Longfei Wang / Di Wu / Carol V Robinson / Hao Wu / Tian-Min Fu /   Abstract: Vesicular- or vacuolar-type adenosine triphosphatases (V-ATPases) are ATP-driven proton pumps comprised of a cytoplasmic V complex for ATP hydrolysis and a membrane-embedded V complex for proton ...Vesicular- or vacuolar-type adenosine triphosphatases (V-ATPases) are ATP-driven proton pumps comprised of a cytoplasmic V complex for ATP hydrolysis and a membrane-embedded V complex for proton transfer. They play important roles in acidification of intracellular vesicles, organelles, and the extracellular milieu in eukaryotes. Here, we report cryoelectron microscopy structures of human V-ATPase in three rotational states at up to 2.9-Å resolution. Aided by mass spectrometry, we build all known protein subunits with associated N-linked glycans and identify glycolipids and phospholipids in the V complex. We define ATP6AP1 as a structural hub for V complex assembly because it connects to multiple V subunits and phospholipids in the c-ring. The glycolipids and the glycosylated V subunits form a luminal glycan coat critical for V-ATPase folding, localization, and stability. This study identifies mechanisms of V-ATPase assembly and biogenesis that rely on the integrated roles of ATP6AP1, glycans, and lipids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21847.map.gz emd_21847.map.gz | 157.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21847-v30.xml emd-21847-v30.xml emd-21847.xml emd-21847.xml | 31.5 KB 31.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21847.png emd_21847.png | 63.3 KB | ||
| Others |  emd_21847_additional_1.map.gz emd_21847_additional_1.map.gz | 168.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21847 http://ftp.pdbj.org/pub/emdb/structures/EMD-21847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21847 | HTTPS FTP |
-Validation report
| Summary document |  emd_21847_validation.pdf.gz emd_21847_validation.pdf.gz | 467.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21847_full_validation.pdf.gz emd_21847_full_validation.pdf.gz | 467 KB | Display | |
| Data in XML |  emd_21847_validation.xml.gz emd_21847_validation.xml.gz | 7.3 KB | Display | |
| Data in CIF |  emd_21847_validation.cif.gz emd_21847_validation.cif.gz | 8.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21847 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21847 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21847 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21847 | HTTPS FTP |
-Related structure data
| Related structure data |  6wm2MC  6wlwC  6wlzC  6wm3C  6wm4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11132 (Title: Cryo-EM structures of human V-ATPase / Data size: 8.4 TB EMPIAR-11132 (Title: Cryo-EM structures of human V-ATPase / Data size: 8.4 TBData #1: Unaligned multi frame micrographs of human V-ATPase in complex with SidK [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21847.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21847.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human V-ATPase in state 1 (composite map) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
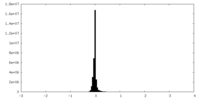
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Human V-ATPase in state 1
| File | emd_21847_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human V-ATPase in state 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
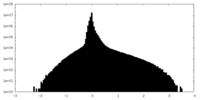
| Density Histograms |
- Sample components
Sample components
+Entire : Human V-ATPase in state 1 with SidK and ADP
+Supramolecule #1: Human V-ATPase in state 1 with SidK and ADP
+Macromolecule #1: V-type proton ATPase subunit E 1
+Macromolecule #2: V-type proton ATPase subunit G 1
+Macromolecule #3: V-type proton ATPase subunit C 1
+Macromolecule #4: V-type proton ATPase subunit H
+Macromolecule #5: V-type proton ATPase 116 kDa subunit a isoform 1
+Macromolecule #6: V-type proton ATPase catalytic subunit A
+Macromolecule #7: V-type proton ATPase subunit B, brain isoform
+Macromolecule #8: SidK
+Macromolecule #9: V-type proton ATPase subunit D
+Macromolecule #10: V-type proton ATPase subunit F
+Macromolecule #11: V-type proton ATPase 21 kDa proteolipid subunit
+Macromolecule #12: V-type proton ATPase 16 kDa proteolipid subunit
+Macromolecule #13: V-type proton ATPase subunit d 1
+Macromolecule #14: V-type proton ATPase subunit e 1
+Macromolecule #15: Ribonuclease kappa
+Macromolecule #16: V-type proton ATPase subunit S1
+Macromolecule #17: Renin receptor
+Macromolecule #22: CHOLESTEROL
+Macromolecule #23: 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE
+Macromolecule #24: PHOSPHATIDYLETHANOLAMINE
+Macromolecule #25: tri(methyl)-[2-[[(2~{R})-2-[(~{Z})-octadec-9-enoyl]oxy-3-[(~{E})-...
+Macromolecule #26: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #27: methyl (3R,6Z,10E,14E)-3,7,11,15,19-pentamethylicosa-6,10,14,18-t...
+Macromolecule #28: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #29: (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1000000 |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller






























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)