[English] 日本語
 Yorodumi
Yorodumi- EMDB-12213: CryoEM structure of disease related M854K MDA5-dsRNA filament in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12213 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of disease related M854K MDA5-dsRNA filament in complex with ATP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PROTEIN-RNA COMPLEX / HELICAL FILAMENT / ATPASE / INNATE IMMUNE RECEPTOR / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationMDA-5 signaling pathway / positive regulation of response to cytokine stimulus / Ub-specific processing proteases / negative regulation of viral genome replication / type I interferon-mediated signaling pathway / cellular response to exogenous dsRNA / pattern recognition receptor activity / protein complex oligomerization / positive regulation of interferon-alpha production / protein sumoylation ...MDA-5 signaling pathway / positive regulation of response to cytokine stimulus / Ub-specific processing proteases / negative regulation of viral genome replication / type I interferon-mediated signaling pathway / cellular response to exogenous dsRNA / pattern recognition receptor activity / protein complex oligomerization / positive regulation of interferon-alpha production / protein sumoylation / ribonucleoprotein complex binding / antiviral innate immune response / positive regulation of interferon-beta production / cellular response to virus / positive regulation of interleukin-6 production / response to virus / positive regulation of tumor necrosis factor production / double-stranded RNA binding / defense response to virus / RNA helicase activity / single-stranded RNA binding / RNA helicase / protein domain specific binding / innate immune response / ATP hydrolysis activity / mitochondrion / DNA binding / zinc ion binding / ATP binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Pseudomonas virus phi6 Pseudomonas virus phi6 | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Singh R / Herrero del Valle A | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA. Authors: Qin Yu / Alba Herrero Del Valle / Rahul Singh / Yorgo Modis /   Abstract: Our innate immune responses to viral RNA are vital defenses. Long cytosolic double-stranded RNA (dsRNA) is recognized by MDA5. The ATPase activity of MDA5 contributes to its dsRNA binding selectivity. ...Our innate immune responses to viral RNA are vital defenses. Long cytosolic double-stranded RNA (dsRNA) is recognized by MDA5. The ATPase activity of MDA5 contributes to its dsRNA binding selectivity. Mutations that reduce RNA selectivity can cause autoinflammatory disease. Here, we show how the disease-associated MDA5 variant M854K perturbs MDA5-dsRNA recognition. M854K MDA5 constitutively activates interferon signaling in the absence of exogenous RNA. M854K MDA5 lacks ATPase activity and binds more stably to synthetic Alu:Alu dsRNA. CryoEM structures of MDA5-dsRNA filaments at different stages of ATP hydrolysis show that the K854 sidechain forms polar bonds that constrain the conformation of MDA5 subdomains, disrupting key steps in the ATPase cycle- RNA footprint expansion and helical twist modulation. The M854K mutation inhibits ATP-dependent RNA proofreading via an allosteric mechanism, allowing MDA5 to form signaling complexes on endogenous RNAs. This work provides insights on how MDA5 recognizes dsRNA in health and disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12213.map.gz emd_12213.map.gz | 8.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12213-v30.xml emd-12213-v30.xml emd-12213.xml emd-12213.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12213_fsc.xml emd_12213_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_12213.png emd_12213.png | 183.5 KB | ||
| Masks |  emd_12213_msk_1.map emd_12213_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12213.cif.gz emd-12213.cif.gz | 8.2 KB | ||
| Others |  emd_12213_half_map_1.map.gz emd_12213_half_map_1.map.gz emd_12213_half_map_2.map.gz emd_12213_half_map_2.map.gz | 65.4 MB 65.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12213 http://ftp.pdbj.org/pub/emdb/structures/EMD-12213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12213 | HTTPS FTP |
-Related structure data
| Related structure data |  7bkpMC  7bkqC  7ngaC  7nicC  7niqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10630 (Title: CryoEM structure of disease related M854K MDA5-dsRNA filament in complex with ATP EMPIAR-10630 (Title: CryoEM structure of disease related M854K MDA5-dsRNA filament in complex with ATPData size: 1.2 TB Data #1: Unaligned multi frame micrographs for the Cryo-EM structure of disease related M854K MDA5-dsRNA filament in complex with ATP [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12213.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12213.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
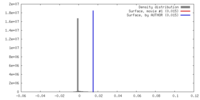
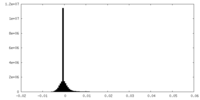
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12213_msk_1.map emd_12213_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
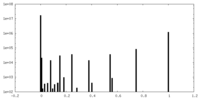
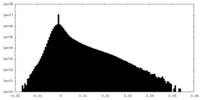
| Density Histograms |
-Half map: #1
| File | emd_12213_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
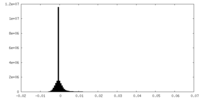
| Density Histograms |
-Half map: #2
| File | emd_12213_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
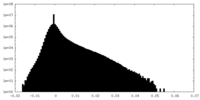
| Density Histograms |
- Sample components
Sample components
-Entire : M854K MDA5-dsRNA filament in complex with ATP
| Entire | Name: M854K MDA5-dsRNA filament in complex with ATP |
|---|---|
| Components |
|
-Supramolecule #1: M854K MDA5-dsRNA filament in complex with ATP
| Supramolecule | Name: M854K MDA5-dsRNA filament in complex with ATP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 1.91 kDa/nm |
-Supramolecule #2: INTERFERON-INDUCED HELICASE C DOMAIN-CONTAINING PROTEIN
| Supramolecule | Name: INTERFERON-INDUCED HELICASE C DOMAIN-CONTAINING PROTEIN type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: RNA
| Supramolecule | Name: RNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Pseudomonas virus phi6 Pseudomonas virus phi6 |
-Macromolecule #1: Interferon-induced helicase C domain-containing protein 1
| Macromolecule | Name: Interferon-induced helicase C domain-containing protein 1 type: protein_or_peptide / ID: 1 / Details: ATP / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 116.122359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSIVCSAEDS FRNLILFFRP RLKMYIQVEP VLDHLIFLSA ETKEQILKKI NTCGNTSAAE LLLSTLEQGQ WPLGWTQMFV EALEHSGNP LAARYVKPTL TDLPSPSSET AHDECLHLLT LLQPTLVDKL LINDVLDTCF EKGLLTVEDR NRISAAGNSG N ESGVRELL ...String: MSIVCSAEDS FRNLILFFRP RLKMYIQVEP VLDHLIFLSA ETKEQILKKI NTCGNTSAAE LLLSTLEQGQ WPLGWTQMFV EALEHSGNP LAARYVKPTL TDLPSPSSET AHDECLHLLT LLQPTLVDKL LINDVLDTCF EKGLLTVEDR NRISAAGNSG N ESGVRELL RRIVQKENWF STFLDVLRQT GNDALFQELT GGGCPEDNTD LANSSHRDGP AANECLLPAV DESSLETEAW NV DDILPEA SCTDSSVTTE SDTSLAEGSV SCFDESLGHN SNMGRDSGTM GSDSDESVIQ TKRVSPEPEL QLRPYQMEVA QPA LDGKNI IICLPTGSGK TRVAVYITKD HLDKKKQASE SGKVIVLVNK VMLAEQLFRK EFNPYLKKWY RIIGLSGDTQ LKIS FPEVV KSYDVIISTA QILENSLLNL ESGDDDGVQL SDFSLIIIDE CHHTNKEAVY NNIMRRYLKQ KLRNNDLKKQ NKPAI PLPQ ILGLTASPGV GAAKKQSEAE KHILNICANL DAFTIKTVKE NLGQLKHQIK EPCKKFVIAD DTRENPFKEK LLEIMA SIQ TYCQKSPMSD FGTQHYEQWA IQMEKKAAKD GNRKDRVCAE HLRKYNEALQ INDTIRMIDA YSHLETFYTD EKEKKFA VL NDSDKSDDEA SSCNDQLKGD VKKSLKLDET DEFLMNLFFD NKKMLKKLAE NPKYENEKLI KLRNTILEQF TRSEESSR G IIFTKTRQST YALSQWIMEN AKFAEVGVKA HHLIGAGHSS EVKPMTQTEQ KEVISKFRTG EINLLIATTV AEEGLDIKE CNIVIRYGLV TNEIAMVQAR GRARADESTY VLVTSSGSGV TEREIVNDFR EKMKYKAINR VQNMKPEEYA HKILELQVQS ILEKKMKVK RSIAKQYNDN PSLITLLCKN CSMLVCSGEN IHVIEKMHHV NMTPEFKGLY IVRENKALQK KFADYQTNGE I ICKCGQAW GTMMVHKGLD LPCLKIRNFV VNFKNNSPKK QYKKWVELPI RFPDLDYSEY CLYSDED UniProtKB: Interferon-induced helicase C domain-containing protein 1 |
-Macromolecule #2: RNA (5'-R(P*CP*UP*CP*UP*CP*CP*UP*CP*GP*GP*CP*UP*UP*G)-3')
| Macromolecule | Name: RNA (5'-R(P*CP*UP*CP*UP*CP*CP*UP*CP*GP*GP*CP*UP*UP*G)-3') type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Pseudomonas virus phi6 Pseudomonas virus phi6 |
| Molecular weight | Theoretical: 4.35258 KDa |
| Sequence | String: CUCUCCUCGG CUUG |
-Macromolecule #3: RNA (5'-R(P*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*AP*G)-3')
| Macromolecule | Name: RNA (5'-R(P*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*AP*G)-3') type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Pseudomonas virus phi6 Pseudomonas virus phi6 |
| Molecular weight | Theoretical: 4.587852 KDa |
| Sequence | String: CAAGCCGAGG AGAG |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: machine step 6 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277.2 K / Instrument: FEI VITROBOT MARK IV Details: Grids were blotted for 3 s; blot force 10, 5, 0, -5, -10. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: OTHER / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 7147 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: -4.0 µm / Calibrated defocus min: -0.25 µm / Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Cs: 2.7 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 50 / Target criteria: Correlation coefficient |
| Output model |  PDB-7bkp: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)