[English] 日本語
 Yorodumi
Yorodumi- EMDB-9888: Structure of influenza D virus polymerase bound to cRNA promoter ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9888 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | |||||||||
 Map data Map data | Structure of influenza D virus polymerase bound to cRNA promoter in mode 2 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / virion component / symbiont-mediated suppression of host gene expression / RNA-directed RNA polymerase / viral RNA genome replication / nucleotide binding / RNA-directed RNA polymerase activity ...cap snatching / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / virion component / symbiont-mediated suppression of host gene expression / RNA-directed RNA polymerase / viral RNA genome replication / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / host cell nucleus / RNA binding Similarity search - Function | |||||||||
| Biological species |  Influenza D virus / synthetic construct (others) Influenza D virus / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Peng Q / Peng R / Qi J / Gao GF / Shi Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2019 Journal: Nat Microbiol / Year: 2019Title: Structural insight into RNA synthesis by influenza D polymerase. Authors: Qi Peng / Yuqian Liu / Ruchao Peng / Min Wang / Wei Yang / Hao Song / Yuhai Chen / Sheng Liu / Min Han / Xinzheng Zhang / Peiyi Wang / Jinghua Yan / Buchang Zhang / Jianxun Qi / Tao Deng / ...Authors: Qi Peng / Yuqian Liu / Ruchao Peng / Min Wang / Wei Yang / Hao Song / Yuhai Chen / Sheng Liu / Min Han / Xinzheng Zhang / Peiyi Wang / Jinghua Yan / Buchang Zhang / Jianxun Qi / Tao Deng / George F Gao / Yi Shi /  Abstract: The influenza virus polymerase uses capped RNA primers to initiate transcription, and a combination of terminal and internal de novo initiations for the two-step replication process by binding the ...The influenza virus polymerase uses capped RNA primers to initiate transcription, and a combination of terminal and internal de novo initiations for the two-step replication process by binding the conserved viral genomic RNA (vRNA) or complementary RNA (cRNA) promoter. Here, we determined the apo and promoter-bound influenza D polymerase structures using cryo-electron microscopy and found the polymerase has an evolutionarily conserved stable core structure with inherently flexible peripheral domains. Strikingly, two conformations (mode A and B) of the vRNA promoter were observed where the 3'-vRNA end can bind at two different sites, whereas the cRNA promoter only binds in the mode B conformation. Functional studies confirmed the critical role of the mode B conformation for vRNA synthesis via the intermediate cRNA but not for cRNA production, which is mainly regulated by the mode A conformation. Both conformations participate in the regulation of the transcription process. This work advances our understanding of the regulatory mechanisms for the synthesis of different RNA species by influenza virus polymerase and opens new opportunities for antiviral drug design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9888.map.gz emd_9888.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9888-v30.xml emd-9888-v30.xml emd-9888.xml emd-9888.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9888_fsc.xml emd_9888_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_9888.png emd_9888.png | 36.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9888 http://ftp.pdbj.org/pub/emdb/structures/EMD-9888 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9888 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9888 | HTTPS FTP |
-Related structure data
| Related structure data |  6kuvMC  9577C  9578C  9579C  9580C  9581C  9582C  9887C  6kujC  6kukC  6kupC  6kurC  6kutC  6kuuC  6kv5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9888.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9888.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of influenza D virus polymerase bound to cRNA promoter in mode 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
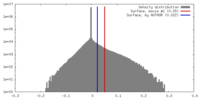
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Influenza D virus polymerase bound to cRNA promoter in mode B con...
| Entire | Name: Influenza D virus polymerase bound to cRNA promoter in mode B conformation |
|---|---|
| Components |
|
-Supramolecule #1: Influenza D virus polymerase bound to cRNA promoter in mode B con...
| Supramolecule | Name: Influenza D virus polymerase bound to cRNA promoter in mode B conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Influenza D virus Influenza D virus |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 270 KDa |
-Macromolecule #1: Polymerase 3
| Macromolecule | Name: Polymerase 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza D virus Influenza D virus |
| Molecular weight | Theoretical: 83.036086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSVIREIAK RFLEQATINI AEEVVREYGD HERTMISVGV HFQACCLISD EYTLEDETTP RYVLLEGLKR QEAISKQNNI CSTLGLEPL RNLADIFDRK TRRFLEVGIT KRESDEYYQE KFNKIGNDMD IHVFTYEGKY FSNNPNGLED IQKTRIFTFL S FVSDELRK ...String: MSSVIREIAK RFLEQATINI AEEVVREYGD HERTMISVGV HFQACCLISD EYTLEDETTP RYVLLEGLKR QEAISKQNNI CSTLGLEPL RNLADIFDRK TRRFLEVGIT KRESDEYYQE KFNKIGNDMD IHVFTYEGKY FSNNPNGLED IQKTRIFTFL S FVSDELRK ENMFTEMYVT EEGAPELEMY KSKLFIAMRD ESVPLPYINY EHLRTRCETF KRNQAECEAK VADVASRLKI KL EHLEENK LRPLEIPKEK EAPYTHKFLM KDAWFFAKPH DSERAQPQQI LYDFFEAANM GFMTTSPKPI FGKQGLMYHS LWG QTKRAI KDKRNELEPS EQRDFLCGIG RASKKIQEDK WQESREEEFK QEETKGAAKR GFPTWFNEEW LWAMRDSGDG DNKI GDWIP MAEMPPCKNE MEDYAKKMCE ELESKIQGTN CAREMSKLIH TIGSLHTECR NFPGKVKIVP IYCRGTLRGE STDCL FGIA IKGKSHLNKD DGMYTVVTFE FSTEEPNPSK HEKYTVFEAG TVPVEAVVLT PKRERVLKEK KLFLYCRTTG MSKLKN DWF SKCRRCLIPT METVEQIVLK ECALKEENRV SEMLENKRAW IAHENGENLT RLVSTKLKDL CRMLIVTQFY YCIYNDN QL EGFCNEQKKF LMFLQADKDS KSAFTFNQKG LYEKIEECIV SNPLCIFLAD RLNKLFLVAK SNGAKYFE |
-Macromolecule #2: RNA-directed RNA polymerase catalytic subunit
| Macromolecule | Name: RNA-directed RNA polymerase catalytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Influenza D virus Influenza D virus |
| Molecular weight | Theoretical: 86.138844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEINPYLLML NNDITSMISL TYPYTGAPPM SHGTSTKYSM ETVSRTYSYS RTKKEVPSGI FPIERRKFCN TIEDKENLEK PNGNVDINF MLSLAEMLEE KMGKGFFKFC ANEAEAEILK MHFSKLTEGR QTYDWTSERN MPAATALQLT VDAIQETQGT F KGTTMVEY ...String: MEINPYLLML NNDITSMISL TYPYTGAPPM SHGTSTKYSM ETVSRTYSYS RTKKEVPSGI FPIERRKFCN TIEDKENLEK PNGNVDINF MLSLAEMLEE KMGKGFFKFC ANEAEAEILK MHFSKLTEGR QTYDWTSERN MPAATALQLT VDAIQETQGT F KGTTMVEY CNKILEMMDW PEVKFKKVRM IVQRHWDPKT KKEIKMKSPT LMITKIGREE FIKRICTINT MAKDGERGKY KR RAIATPG MGIRPFSKIV ETLAQKICER LAESGLPVGG NEKKAKLKTT VSSTNSKLQE GQFMVNITGD NSKWNECQQP EAY LAMLAY ITKDSSNLMK DLCSVAPTLF CNKYVKMGQG FRAKNKRKTK EIVIPAKKMK ERKELMNAEW RDLFETIEPY MDGE CCFLG GGMLMGMFNM LSTVFGVMTL NYREEALARR NCYWTGLQSS DDFVLFCISR TWPEMEMTIL KFIAVCKLMG INMSL EKSY GCLPELFEFT SMFFSGDFVS NIALELPAFT TAGMNEGTDF TAAMSVIRTN MINNGLSPGT ALMALRICLQ EFRATY RVH PYDSGVKNHR MKIIRKFIET IENKDGLLIS DGGKLMNNIS SLHIPEEILK EDLMDPSYRN RVFNPRNPFT QFEKTVD IF KASGPIRVEE NEAVVSTHSF RTRSNRTLLN TDMRAMALEE KRYQVVCNMY RSVFESADVN TPIGSMSMGE AIEAKILD R ARTQFENGII GGEEYSEIKR LIEDAKRQRL SV |
-Macromolecule #3: Polymerase PB2
| Macromolecule | Name: Polymerase PB2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza D virus Influenza D virus |
| Molecular weight | Theoretical: 88.480484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLLLTLAKE YANLTKDKKS CKLLSQGTVS SYTTFKKWTT SRKEKNPSLR MRWAMGSKFP IMANREILEE AGIPEQWEGI DLWSKKDDV SKLGMVLASP AAITYWNFCG PGVDNSSVIK DVYKAKFMKK ERWRETLWGP MNFELVGKQR RVVETQPVEI K LNQKEIKE ...String: MSLLLTLAKE YANLTKDKKS CKLLSQGTVS SYTTFKKWTT SRKEKNPSLR MRWAMGSKFP IMANREILEE AGIPEQWEGI DLWSKKDDV SKLGMVLASP AAITYWNFCG PGVDNSSVIK DVYKAKFMKK ERWRETLWGP MNFELVGKQR RVVETQPVEI K LNQKEIKE LTMWVLFEDE ANLASKFIQE NFSLVLSLRE LYKGKAVNKD VAAFMIAHQF SPEKRFLPTF GPIRPERMEL LH CLGGDFW KIEAVTAGSL NEEQKKRDVR AVARKICLRA SVDLFTPAEK IRDYIASVTM RFGTVERTFE DVIRNSDDIS AEV TLCKAA LGCELGKSMS FGNLNLRKVS GEAETMEKTV YWGLKPIKYK CWRGEETFYC ELRKVTCMFR RSEGLDWANI GPGS PEERR ELLAMVMIFC RDGRFFESAP VNIDESFFRT RLNKEIPYQY VLLKWVRQSR DNLDALLSTR GLIPAHIGQF GKGMG IDGS SSSSMVYKGV MLSKTPIDIV ESKEKHRLFL NDNIEAVTER GAMVASIMDL SEDNRETFND VTFNHVDLAV LKDEKT AII KIYRSLVERI NTDDDGLPAL IMGKRYLELY QLDEVKDAVG LIPKRMLGAY SYQARQLIQS QIKNDSYSLP EIIKLLP FC YSPPKKMLFD GTFHFKNQMY VRPGINTNLF SFSKTDKSKI YVNGSAVKIK LVLGDDEMDT SLAFVEGFQV CEYDPRAP L IPRRDLRLIG FGKKVRVFVG QGQEKTLVRT SSKRAASHDV SKNIRRMRLE V |
-Macromolecule #4: RNA (5'-R(P*UP*CP*CP*UP*UP*GP*CP*UP*AP*CP*UP*GP*CP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*CP*CP*UP*UP*GP*CP*UP*AP*CP*UP*GP*CP*U)-3') type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.337563 KDa |
| Sequence | String: UCCUUGCUAC UGCU |
-Macromolecule #5: RNA (5'-R(P*AP*GP*CP*AP*UP*AP*AP*GP*CP*AP*GP*GP*AP*GP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*GP*CP*AP*UP*AP*AP*GP*CP*AP*GP*GP*AP*GP*A)-3') type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.902042 KDa |
| Sequence | String: AGCAUAAGCA GGAGA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6kuv: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)