[English] 日本語
 Yorodumi
Yorodumi- EMDB-9192: Cryo-electron microscopy structure of Plasmodium falciparum Rh5/C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9192 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy structure of Plasmodium falciparum Rh5/CyRPA/Ripr invasion complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Invasion Ligand Protein complex / CELL INVASION | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicroneme lumen / microneme / symbiont entry into host / apical part of cell / cytoplasmic vesicle / host extracellular space / host cell plasma membrane / protein-containing complex / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |    | |||||||||
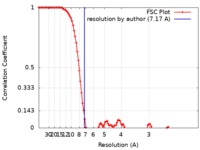
| Method | single particle reconstruction / cryo EM / Resolution: 7.17 Å | |||||||||
 Authors Authors | Wilson W / Zhiheng Y | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structure of Plasmodium falciparum Rh5-CyRPA-Ripr invasion complex. Authors: Wilson Wong / Rick Huang / Sebastien Menant / Chuan Hong / Jarrod J Sandow / Richard W Birkinshaw / Julie Healer / Anthony N Hodder / Usheer Kanjee / Christopher J Tonkin / Denise Heckmann / ...Authors: Wilson Wong / Rick Huang / Sebastien Menant / Chuan Hong / Jarrod J Sandow / Richard W Birkinshaw / Julie Healer / Anthony N Hodder / Usheer Kanjee / Christopher J Tonkin / Denise Heckmann / Vladislav Soroka / Teit Max Moscote Søgaard / Thomas Jørgensen / Manoj T Duraisingh / Peter E Czabotar / Willem A de Jongh / Wai-Hong Tham / Andrew I Webb / Zhiheng Yu / Alan F Cowman /    Abstract: Plasmodium falciparum causes the severe form of malaria that has high levels of mortality in humans. Blood-stage merozoites of P. falciparum invade erythrocytes, and this requires interactions ...Plasmodium falciparum causes the severe form of malaria that has high levels of mortality in humans. Blood-stage merozoites of P. falciparum invade erythrocytes, and this requires interactions between multiple ligands from the parasite and receptors in hosts. These interactions include the binding of the Rh5-CyRPA-Ripr complex with the erythrocyte receptor basigin, which is an essential step for entry into human erythrocytes. Here we show that the Rh5-CyRPA-Ripr complex binds the erythrocyte cell line JK-1 significantly better than does Rh5 alone, and that this binding occurs through the insertion of Rh5 and Ripr into host membranes as a complex with high molecular weight. We report a cryo-electron microscopy structure of the Rh5-CyRPA-Ripr complex at subnanometre resolution, which reveals the organization of this essential invasion complex and the mode of interactions between members of the complex, and shows that CyRPA is a critical mediator of complex assembly. Our structure identifies blades 4-6 of the β-propeller of CyRPA as contact sites for Rh5 and Ripr. The limited contacts between Rh5-CyRPA and CyRPA-Ripr are consistent with the dissociation of Rh5 and Ripr from CyRPA for membrane insertion. A comparision of the crystal structure of Rh5-basigin with the cryo-electron microscopy structure of Rh5-CyRPA-Ripr suggests that Rh5 and Ripr are positioned parallel to the erythrocyte membrane before membrane insertion. This provides information on the function of this complex, and thereby provides insights into invasion by P. falciparum. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9192.map.gz emd_9192.map.gz | 62.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9192-v30.xml emd-9192-v30.xml emd-9192.xml emd-9192.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9192_fsc.xml emd_9192_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_9192.png emd_9192.png | 86.4 KB | ||
| Filedesc metadata |  emd-9192.cif.gz emd-9192.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9192 http://ftp.pdbj.org/pub/emdb/structures/EMD-9192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9192 | HTTPS FTP |
-Related structure data
| Related structure data |  6mpvMC  9193C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9192.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9192.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PfRh5/CyRPA/PfRipr complex
| Entire | Name: PfRh5/CyRPA/PfRipr complex |
|---|---|
| Components |
|
-Supramolecule #1: PfRh5/CyRPA/PfRipr complex
| Supramolecule | Name: PfRh5/CyRPA/PfRipr complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cysteine-rich protective antigen
| Macromolecule | Name: Cysteine-rich protective antigen / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.270195 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: RHVFIRTELS FIKNNVPCIR DMFFIYKREL YNICLDDLKG EEDETHIYVQ KKVKDSWITL NDLFKETDLT GRPHIFAYVD VEEIIILLC EDEEFSNRKK DMTCHRFYSN DGKEYNNAEI TISDYILKDK LLSSYVSLPL KIENREYFLI CGVSPYKFKD D NKKDDILC ...String: RHVFIRTELS FIKNNVPCIR DMFFIYKREL YNICLDDLKG EEDETHIYVQ KKVKDSWITL NDLFKETDLT GRPHIFAYVD VEEIIILLC EDEEFSNRKK DMTCHRFYSN DGKEYNNAEI TISDYILKDK LLSSYVSLPL KIENREYFLI CGVSPYKFKD D NKKDDILC MASHDKGETW GTKIVIKYDN YKLGVQYFFL RPYISKNDLS FHFYVGDNIN NVKNVNFIEC THEKDLEFVC SN RDFLKDN KVLQDVSTLN DEYIVSYGND NNFAECYIFF NNENSILIKP EKYGNTTAGC YGGTFVKIDE NRALFIYSSS QGI YNIHTI YYANYE UniProtKB: Cysteine-rich protective antigen |
-Macromolecule #2: Reticulocyte binding protein 5
| Macromolecule | Name: Reticulocyte binding protein 5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.803371 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: IIPHYTFLDY YKHLSYNSIY HKSSTYGKCI AVDAFIKKIN ETYDKVKSKC NDIKNDLIAT IKKLEHPYDI NNKNDDSYRY DISEEIDDK SEETDDETEE VEDSIQDTDS NHTPSNKKKN DLMNRTFKKM MDEYNTKKKK LIKCIKNHEN DFNKICMDMK N YGTNLFEQ ...String: IIPHYTFLDY YKHLSYNSIY HKSSTYGKCI AVDAFIKKIN ETYDKVKSKC NDIKNDLIAT IKKLEHPYDI NNKNDDSYRY DISEEIDDK SEETDDETEE VEDSIQDTDS NHTPSNKKKN DLMNRTFKKM MDEYNTKKKK LIKCIKNHEN DFNKICMDMK N YGTNLFEQ LSCYNNNFCN TNGIRYHYDE YIHKLILSVK SKNLNKDLSD MTNILQQSEL LLTNLNKKMG SYIYIDTIKF IH KEMKHIF NRIEYHTKII NDKTKIIQDK IKLNIWRTFQ KDELLKRILD MSNEYSLFIT SDHLRQMLYN TFYSKEKHLN NIF HHLIYV LQMK UniProtKB: Reticulocyte binding protein 5 |
-Macromolecule #3: PfRipr
| Macromolecule | Name: PfRipr / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.379692 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #4: PfRipr
| Macromolecule | Name: PfRipr / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 443.539 Da |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 / Details: 20 mM Tris, pH 8.5, 150 mM NaCl |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Microscope was equipped with a Cs corrector with two hexapole elements Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 4 / Number real images: 12974 / Average exposure time: 10.0 sec. / Average electron dose: 92.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 48077 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)