+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8734 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of "immature" Chikungunya VLP | |||||||||
 Map data Map data | Chikungunya virus strain Senegal 37997 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Chikungunya / virus / immature / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / host cell endoplasmic reticulum / channel activity / monoatomic ion transmembrane transport / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell Golgi apparatus / entry receptor-mediated virion attachment to host cell / host cell cytoplasm / serine-type endopeptidase activity ...togavirin / T=4 icosahedral viral capsid / host cell endoplasmic reticulum / channel activity / monoatomic ion transmembrane transport / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell Golgi apparatus / entry receptor-mediated virion attachment to host cell / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Chikungunya virus strain Senegal 37997 / Chikungunya virus strain Senegal 37997 /  Chikungunya virus (strain 37997) Chikungunya virus (strain 37997) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.8 Å | |||||||||
 Authors Authors | Rossmann MG / Yap ML / Sakurai A / Ishikawa M / Akahata W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Structural studies of Chikungunya virus maturation. Authors: Moh Lan Yap / Thomas Klose / Akane Urakami / S Saif Hasan / Wataru Akahata / Michael G Rossmann /   Abstract: Cleavage of the alphavirus precursor glycoprotein p62 into the E2 and E3 glycoproteins before assembly with the nucleocapsid is the key to producing fusion-competent mature spikes on alphaviruses. ...Cleavage of the alphavirus precursor glycoprotein p62 into the E2 and E3 glycoproteins before assembly with the nucleocapsid is the key to producing fusion-competent mature spikes on alphaviruses. Here we present a cryo-EM, 6.8-Å resolution structure of an "immature" Chikungunya virus in which the cleavage site has been mutated to inhibit proteolysis. The spikes in the immature virus have a larger radius and are less compact than in the mature virus. Furthermore, domains B on the E2 glycoproteins have less freedom of movement in the immature virus, keeping the fusion loops protected under domain B. In addition, the nucleocapsid of the immature virus is more compact than in the mature virus, protecting a conserved ribosome-binding site in the capsid protein from exposure. These differences suggest that the posttranslational processing of the spikes and nucleocapsid is necessary to produce infectious virus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8734.map.gz emd_8734.map.gz | 249 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8734-v30.xml emd-8734-v30.xml emd-8734.xml emd-8734.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
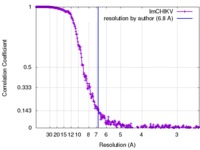
| FSC (resolution estimation) |  emd_8734_fsc.xml emd_8734_fsc.xml | 27.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_8734.png emd_8734.png | 270.3 KB | ||
| Filedesc metadata |  emd-8734.cif.gz emd-8734.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8734 http://ftp.pdbj.org/pub/emdb/structures/EMD-8734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8734 | HTTPS FTP |
-Related structure data
| Related structure data |  5vu2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8734.map.gz / Format: CCP4 / Size: 1.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8734.map.gz / Format: CCP4 / Size: 1.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chikungunya virus strain Senegal 37997 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Chikungunya virus strain Senegal 37997
| Entire | Name:  Chikungunya virus strain Senegal 37997 Chikungunya virus strain Senegal 37997 |
|---|---|
| Components |
|
-Supramolecule #1: Chikungunya virus strain Senegal 37997
| Supramolecule | Name: Chikungunya virus strain Senegal 37997 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 371095 / Sci species name: Chikungunya virus strain Senegal 37997 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: E1 envelope glycoprotein
| Macromolecule | Name: E1 envelope glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain Senegal 37997 / Strain: 37997 Chikungunya virus strain Senegal 37997 / Strain: 37997 |
| Molecular weight | Theoretical: 95.538164 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STKDNFNVYK ATRPYLAHCP DCGEGHSCHS PVALERIRNE ATDGTLKIQV SLQIGIKTDD SHDWTKLRYM DNHMPADAER AGLFVRTSA PCTITGTMGH FILARCPKGE TLTVGFTDSR KISHSCTHPF HHDPPVIGRE KFHSRPQHGK ELPCSTYVQS T AATTEEIE ...String: STKDNFNVYK ATRPYLAHCP DCGEGHSCHS PVALERIRNE ATDGTLKIQV SLQIGIKTDD SHDWTKLRYM DNHMPADAER AGLFVRTSA PCTITGTMGH FILARCPKGE TLTVGFTDSR KISHSCTHPF HHDPPVIGRE KFHSRPQHGK ELPCSTYVQS T AATTEEIE VHMPPDTPDR TLMSQQSGNV KITVNGQTVR YKCNCGGSNE GLTTTDKVIN NCKVDQCHAA VTNHKKWQYN SP LVPRNAE LGDRKGKIHI PFPLANVTCR VPKARNPTVT YGKNQVIMLL YPDHPTLLSY RNMGEEPNYQ EEWVMHKKEV VLT VPTEGL EVTWGNNEPY KYWPQLSTNG TAHGHPHEII LYYYELYPTM TVVVVSVATF ILLSMVGMAA GMCMCARRRC ITPY ELTPG ATVPFLLSLI CCIRTAKAAT YQEAAIYLWN EQQPLFWLQA LIPLAALIVL CNCLRLLPCC CKTLAFLAVM SVGAH TVSA YEHVTVIPNT VGVPYKTLVN RPGYSPMVLE MELLSVTLEP TLSLDYITCE YKTVIPSPYV KCCGTAECKD KNLPDY SCK VFTGVYPFMW GGAYCFCDAE NTQLSEAHVE KSESCKTEFA SAYRAHTASA SAKLRVLYQG NNITVTAYAN GDHAVTV KD AKFIVGPMSS AWTPFDNKIV VYKGDVYNMD YPPFGAGRPG QFGDIQSRTP ESKDVYANTQ LVLQRPAAGT VHVPYSQA P SGFKYWLKER GASLQHTAPF GCQIATNPVR AVNCAVGNMP ISIDIPEAAF TRVVDAPSLT DMSCEVPACT HSSDFGGVA IIKYAASKKG KCAVHSMTNA VTIREAEIEV EGNSQLQISF STALASAEFR VQVCSTQVHC AAECHP UniProtKB: Structural polyprotein |
-Macromolecule #2: E3 envelope glycoprotein
| Macromolecule | Name: E3 envelope glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain Senegal 37997 / Strain: 37997 Chikungunya virus strain Senegal 37997 / Strain: 37997 |
| Molecular weight | Theoretical: 6.92699 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PVMCLLANTT FPCSQPPCTP CCYEKEPEET LRMLEDNVMR PGYYQLLQAS LTCSPHRQRE UniProtKB: Structural polyprotein |
-Macromolecule #3: capsid protein
| Macromolecule | Name: capsid protein / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus (strain 37997) / Strain: 37997 Chikungunya virus (strain 37997) / Strain: 37997 |
| Molecular weight | Theoretical: 16.229508 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CIFEVKHEGK VMGYACLVGD KVMKPAHVKG TIDNADLAKL AFKRSSKYDL ECAQIPVHMK SDASKFTHEK PEGYYNWHHG AVQYSGGRF TIPTGAGKPG DSGRPIFDNK GRVVAIVLGG ANEGARTALS VVTWNKDIVT KITPEGAEEW UniProtKB: Structural polyprotein |
-Macromolecule #4: E1 envelope glycoprotein
| Macromolecule | Name: E1 envelope glycoprotein / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain Senegal 37997 / Strain: 37997 Chikungunya virus strain Senegal 37997 / Strain: 37997 |
| Molecular weight | Theoretical: 4.810723 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: HTTLGVQDIS TTAMSWVQKI TGGVGLIVAV AALILIVVLC VSFSRH UniProtKB: Structural polyprotein |
-Macromolecule #5: E2 envelope glycoprotein
| Macromolecule | Name: E2 envelope glycoprotein / type: protein_or_peptide / ID: 5 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain Senegal 37997 / Strain: 37997 Chikungunya virus strain Senegal 37997 / Strain: 37997 |
| Molecular weight | Theoretical: 8.817458 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STNGTAHGHP HEIILYYYEL YPTMTVVIVS VASFVLLSMV GTAVGMCVCA RRRCITPYEL TPGATVPFLL SLLCCVRTTK A UniProtKB: Structural polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 36.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)