+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8669 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map of MsbA in POPG nanodisc | |||||||||
 Map data Map data | map of MsbA with LPS in POPG nanodisc, filtered to 5A and sharpened with -280 B-factor | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type lipid A-core oligosaccharide transporter / ATPase-coupled lipid transmembrane transporter activity / ABC-type transporter activity / ATP hydrolysis activity / ATP binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
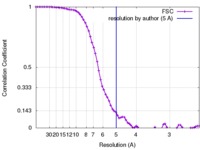
| Method | single particle reconstruction / cryo EM / Resolution: 5.0 Å | |||||||||
 Authors Authors | Mi W / Walz T / Liao M | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Structural basis of MsbA-mediated lipopolysaccharide transport. Authors: Wei Mi / Yanyan Li / Sung Hwan Yoon / Robert K Ernst / Thomas Walz / Maofu Liao /   Abstract: Lipopolysaccharide (LPS) in the outer membrane of Gram-negative bacteria is critical for the assembly of their cell envelopes. LPS synthesized in the cytoplasmic leaflet of the inner membrane is ...Lipopolysaccharide (LPS) in the outer membrane of Gram-negative bacteria is critical for the assembly of their cell envelopes. LPS synthesized in the cytoplasmic leaflet of the inner membrane is flipped to the periplasmic leaflet by MsbA, an ATP-binding cassette transporter. Despite substantial efforts, the structural mechanisms underlying MsbA-driven LPS flipping remain elusive. Here we use single-particle cryo-electron microscopy to elucidate the structures of lipid-nanodisc-embedded MsbA in three functional states. The 4.2 Å-resolution structure of the transmembrane domains of nucleotide-free MsbA reveals that LPS binds deep inside MsbA at the height of the periplasmic leaflet, establishing extensive hydrophilic and hydrophobic interactions with MsbA. Two sub-nanometre-resolution structures of MsbA with ADP-vanadate and ADP reveal an unprecedented closed and an inward-facing conformation, respectively. Our study uncovers the structural basis for LPS recognition, delineates the conformational transitions of MsbA to flip LPS, and paves the way for structural characterization of other lipid flippases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8669.map.gz emd_8669.map.gz | 24.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8669-v30.xml emd-8669-v30.xml emd-8669.xml emd-8669.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8669_fsc.xml emd_8669_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_8669.png emd_8669.png | 55.3 KB | ||
| Others |  emd_8669_additional.map.gz emd_8669_additional.map.gz | 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8669 http://ftp.pdbj.org/pub/emdb/structures/EMD-8669 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8669 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8669 | HTTPS FTP |
-Validation report
| Summary document |  emd_8669_validation.pdf.gz emd_8669_validation.pdf.gz | 78.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8669_full_validation.pdf.gz emd_8669_full_validation.pdf.gz | 77.8 KB | Display | |
| Data in XML |  emd_8669_validation.xml.gz emd_8669_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8669 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8669 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8669 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8669 | HTTPS FTP |
-Related structure data
| Related structure data |  8465C  8467C  8469C  8670C  8671C  5ttpC  5tv4C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8669.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8669.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of MsbA with LPS in POPG nanodisc, filtered to 5A and sharpened with -280 B-factor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.28 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: map without any filter or bfactor sharpen
| File | emd_8669_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map without any filter or bfactor sharpen | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MsbA with LPS in POPG nanodisc
| Entire | Name: MsbA with LPS in POPG nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: MsbA with LPS in POPG nanodisc
| Supramolecule | Name: MsbA with LPS in POPG nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 67.2 KDa |
-Macromolecule #1: MsbA
| Macromolecule | Name: MsbA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH HHSSGHIDDD DKHMHNDKDL STWQTFRRLW PTIAPFKAGL IVAGVALILN AASDTFMLSL LKPLLDDGFG KTDRSVLVWM PLVVIGLMIL RGITSYVSSY CISWVSGKVV MTMRRRLFGH MMGMPVSFFD KQSTGTLLSR ITYDSEQVAS SSSGALITVV ...String: MGHHHHHHHH HHSSGHIDDD DKHMHNDKDL STWQTFRRLW PTIAPFKAGL IVAGVALILN AASDTFMLSL LKPLLDDGFG KTDRSVLVWM PLVVIGLMIL RGITSYVSSY CISWVSGKVV MTMRRRLFGH MMGMPVSFFD KQSTGTLLSR ITYDSEQVAS SSSGALITVV REGASIIGLF IMMFYYSWQL SIILIVLAPI VSIAIRVVSK RFRNISKNMQ NTMGQVTTSA EQMLKGHKEV LIFGGQEVET KRFDKVSNRM RLQGMKMVSA SSISDPIIQL IASLALAFVL YAASFPSVMD SLTAGTITVV FSSMIALMRP LKSLTNVNAQ FQRGMAACQT LFTILDSEQE KDEGKRVIER ATGDVEFRNV TFTYPGRDVP ALRNINLKIP AGKTVALVGR SGSGKSTIAS LITRFYDIDE GEILMDGHDL REYTLASLRN QVALVSQNVH LFNDTVANNI AYARTEQYSR EQIEEAARMA YAMDFINKMD NGLDTVIGEN GVLLSGGQRQ RIAIARALLR DSPILILDEA TSALDTESER AIQAALDELQ KNRTSLVIAH RLSTIEKADE IVVVEDGVIV ERGTHNDLLE HRGVYAQLHK MQFGQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 7.2 sec. / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X