[English] 日本語
 Yorodumi
Yorodumi- EMDB-7330: High-Resolution Cryo-EM Structures of Actin-bound Myosin States R... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7330 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | |||||||||
 Map data Map data | Actin-bound Myosin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mechanochemistry / Mechanobiology / Structural Biology / Cytoskeleton / Molecular Motor / Myosin-I / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpost-Golgi vesicle-mediated transport / actin filament-based movement / myosin complex / CaM pathway / Cam-PDE 1 activation / Sodium/Calcium exchangers / Calmodulin induced events / cytoskeletal motor activator activity / Reduction of cytosolic Ca++ levels / Activation of Ca-permeable Kainate Receptor ...post-Golgi vesicle-mediated transport / actin filament-based movement / myosin complex / CaM pathway / Cam-PDE 1 activation / Sodium/Calcium exchangers / Calmodulin induced events / cytoskeletal motor activator activity / Reduction of cytosolic Ca++ levels / Activation of Ca-permeable Kainate Receptor / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde / Loss of phosphorylation of MECP2 at T308 / CREB1 phosphorylation through the activation of Adenylate Cyclase / negative regulation of high voltage-gated calcium channel activity / PKA activation / CaMK IV-mediated phosphorylation of CREB / microfilament motor activity / Glycogen breakdown (glycogenolysis) / myosin heavy chain binding / CLEC7A (Dectin-1) induces NFAT activation / Activation of RAC1 downstream of NMDARs / negative regulation of ryanodine-sensitive calcium-release channel activity / organelle localization by membrane tethering / mitochondrion-endoplasmic reticulum membrane tethering / autophagosome membrane docking / tropomyosin binding / negative regulation of calcium ion export across plasma membrane / regulation of cardiac muscle cell action potential / presynaptic endocytosis / actin filament bundle / troponin I binding / filamentous actin / microvillus / regulation of cell communication by electrical coupling involved in cardiac conduction / Synthesis of IP3 and IP4 in the cytosol / mesenchyme migration / phosphatidylinositol-3,4,5-trisphosphate binding / Phase 0 - rapid depolarisation / Negative regulation of NMDA receptor-mediated neuronal transmission / calcineurin-mediated signaling / Unblocking of NMDA receptors, glutamate binding and activation / RHO GTPases activate PAKs / cytoskeletal motor activity / skeletal muscle myofibril / regulation of ryanodine-sensitive calcium-release channel activity / brush border / actin filament bundle assembly / Ion transport by P-type ATPases / striated muscle thin filament / Uptake and function of anthrax toxins / skeletal muscle thin filament assembly / Long-term potentiation / protein phosphatase activator activity / actin monomer binding / Calcineurin activates NFAT / Regulation of MECP2 expression and activity / DARPP-32 events / Smooth Muscle Contraction / detection of calcium ion / regulation of cardiac muscle contraction / catalytic complex / RHO GTPases activate IQGAPs / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / calcium channel inhibitor activity / Activation of AMPK downstream of NMDARs / presynaptic cytosol / skeletal muscle fiber development / cellular response to interferon-beta / stress fiber / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / Protein methylation / Ion homeostasis / eNOS activation / titin binding / Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation / phosphatidylinositol-4,5-bisphosphate binding / regulation of calcium-mediated signaling / actin filament polymerization / voltage-gated potassium channel complex / FCERI mediated Ca+2 mobilization / calcium channel complex / substantia nigra development / regulation of heart rate / FCGR3A-mediated IL10 synthesis / Ras activation upon Ca2+ influx through NMDA receptor / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / actin filament organization / calyx of Held / adenylate cyclase activator activity / protein serine/threonine kinase activator activity / VEGFR2 mediated cell proliferation / trans-Golgi network membrane / sarcomere / regulation of cytokinesis / VEGFR2 mediated vascular permeability / cell periphery / spindle microtubule / positive regulation of receptor signaling pathway via JAK-STAT / Translocation of SLC2A4 (GLUT4) to the plasma membrane / actin filament Similarity search - Function | |||||||||
| Biological species | unidentified (others) /   | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Mentes A / Huehn A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Authors: Ahmet Mentes / Andrew Huehn / Xueqi Liu / Adam Zwolak / Roberto Dominguez / Henry Shuman / E Michael Ostap / Charles V Sindelar /  Abstract: Myosins adjust their power outputs in response to mechanical loads in an isoform-dependent manner, resulting in their ability to dynamically adapt to a range of motile challenges. Here, we reveal the ...Myosins adjust their power outputs in response to mechanical loads in an isoform-dependent manner, resulting in their ability to dynamically adapt to a range of motile challenges. Here, we reveal the structural basis for force-sensing based on near-atomic resolution structures of one rigor and two ADP-bound states of myosin-IB (myo1b) bound to actin, determined by cryo-electron microscopy. The two ADP-bound states are separated by a 25° rotation of the lever. The lever of the first ADP state is rotated toward the pointed end of the actin filament and forms a previously unidentified interface with the N-terminal subdomain, which constitutes the upper half of the nucleotide-binding cleft. This pointed-end orientation of the lever blocks ADP release by preventing the N-terminal subdomain from the pivoting required to open the nucleotide binding site, thus revealing how myo1b is inhibited by mechanical loads that restrain lever rotation. The lever of the second ADP state adopts a rigor-like orientation, stabilized by class-specific elements of myo1b. We identify a role for this conformation as an intermediate in the ADP release pathway. Moreover, comparison of our structures with other myosins reveals structural diversity in the actomyosin binding site, and we reveal the high-resolution structure of actin-bound phalloidin, a potent stabilizer of filamentous actin. These results provide a framework to understand the spectrum of force-sensing capacities among the myosin superfamily. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7330.map.gz emd_7330.map.gz | 194.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7330-v30.xml emd-7330-v30.xml emd-7330.xml emd-7330.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7330.png emd_7330.png | 163.8 KB | ||
| Masks |  emd_7330_msk_1.map emd_7330_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-7330.cif.gz emd-7330.cif.gz | 6.2 KB | ||
| Others |  emd_7330_half_map_1.map.gz emd_7330_half_map_1.map.gz emd_7330_half_map_2.map.gz emd_7330_half_map_2.map.gz | 194.2 MB 194.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7330 http://ftp.pdbj.org/pub/emdb/structures/EMD-7330 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7330 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7330 | HTTPS FTP |
-Related structure data
| Related structure data |  6c1gMC  7329C  7331C  5v7xC  6c1dC  6c1hC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7330.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7330.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin-bound Myosin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
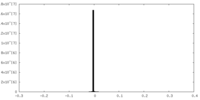
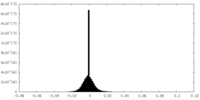
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7330_msk_1.map emd_7330_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
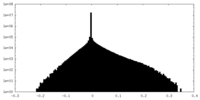
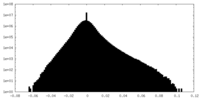
| Density Histograms |
-Half map: Actin-bound Myosin, half mask 1
| File | emd_7330_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin-bound Myosin, half mask 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
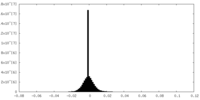
| Density Histograms |
-Half map: Actin-bound Myosin, half mask 2
| File | emd_7330_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin-bound Myosin, half mask 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
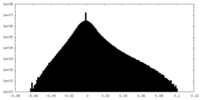
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of actin, myosin-1b, and calmodulin with ADP
| Entire | Name: Complex of actin, myosin-1b, and calmodulin with ADP |
|---|---|
| Components |
|
-Supramolecule #1: Complex of actin, myosin-1b, and calmodulin with ADP
| Supramolecule | Name: Complex of actin, myosin-1b, and calmodulin with ADP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Macromolecule #1: Unconventional myosin-Ib
| Macromolecule | Name: Unconventional myosin-Ib / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 84.14393 KDa |
| Sequence | String: VKSSLLDNMI GVGDTVLLEP LNEETFIDNL KKRFDHNEIY TYIGSVVISV NPYRSLPIYS PEKVEDYRNR NFYELSPHIF ALSDEAYRS LRDQDKDQCI LITGESGAGK TEASKLVMSY VAAVCGKGAE VNQVKEQLLQ STPVLEAFGN AKTVRNDNSS R FGKYMDIE ...String: VKSSLLDNMI GVGDTVLLEP LNEETFIDNL KKRFDHNEIY TYIGSVVISV NPYRSLPIYS PEKVEDYRNR NFYELSPHIF ALSDEAYRS LRDQDKDQCI LITGESGAGK TEASKLVMSY VAAVCGKGAE VNQVKEQLLQ STPVLEAFGN AKTVRNDNSS R FGKYMDIE FDFKGDPLGG VISNYLLEKS RVVKQPRGER NFHVFYQLLS GASEELLHKL KLERDFSRYN YLSLDSAKVN GV DDAANFR TVRNAMQIVG FSDPEAESVL EVVAAVLKLG NIEFKPESRM NGLDESKIKD KNELKEICEL TSIDQVVLER AFS FRTVEA KQEKVSTTLN VAQAYYARDA LAKNLYSRLF SWLVNRINES IKAQTKVRKK VMGVLDIYGF EIFEDNSFEQ FIIN YCNEK LQQIFIELTL KEEQEEYIRE DIEWTHIDYF NNAIICDLIE NNTNGILAML DEECLRPGTV TDETFLEKLN QVCAT HQHF ESRMSKCSRF LNDTTLPHSC FRIQHYAGKV LYQVEGFVDK NNDLLYRDLS QAMWKAGHAL IKSLFPEGNP AKVNLK RPP TAGSQFKASV ATLMKNLQTK NPNYIRCIKP NDKKAAHIFS ESLVCHQIRY LGLLENVRVR RAGYAFRQAY EPCLERY KM LCKQTWPHWK GPARSGVEVL FNELEIPVEE YSFGRSKIFI RNPRTLFQLE DLRKQRLEDL ATLIQKIYRG WKCRTHFL L MKGLNDIF UniProtKB: Unconventional myosin-Ib |
-Macromolecule #2: Calmodulin
| Macromolecule | Name: Calmodulin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Molecular weight | Theoretical: 16.72135 KDa |
| Sequence | String: ADQLTEEQIA EFKEAFSLFD KDGDGTITTK ELGTVMRSLG QNPTEAELQD MINEVDADGN GTIDFPEFLT MMARKMKDTD SEEEIREAF RVFDKDGNGY ISAAELRHVM TNLGEKLTDE EVDEMIREAD IDGDGQVNYE EFVQMMTAK |
-Macromolecule #3: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 3 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.862613 KDa |
| Sequence | String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIITNWD DMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSGDGV T HNVPIYEG ...String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIITNWD DMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSGDGV T HNVPIYEG YALPHAIMRL DLAGRDLTDY LMKILTERGY SFVTTAEREI VRDIKEKLCY VALDFENEMA TAASSSSLEK SY ELPDGQV ITIGNERFRC PETLFQPSFI GMESAGIHET TYNSIMKCDI DIRKDLYANN VMSGGTTMYP GIADRMQKEI TAL APSTMK IKIIAPPERK YSVWIGGSIL ASLSTFQQMW ITKQEYDEAG PSIVHRKCF UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 11.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 27.5 Å Applied symmetry - Helical parameters - Δ&Phi: -167.4 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF Details: Resolution estimated by post-processing in RELION using a mask with soft edges that included only the central subunit. Number images used: 7700 |
|---|---|
| Startup model | Type of model: OTHER |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6c1g: |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)