[English] 日本語
 Yorodumi
Yorodumi- PDB-6zrq: two-protofilament amyloid structure of S20G variant of human amyl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6zrq | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
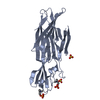
| Title | two-protofilament amyloid structure of S20G variant of human amylin (IAPP - islet amyloid polypeptide) | |||||||||
 Components Components | Islet amyloid polypeptide | |||||||||
 Keywords Keywords | PROTEIN FIBRIL / amyloid fibril type-2-diabetes early-onset | |||||||||
| Function / homology |  Function and homology information Function and homology informationamylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption / eating behavior / positive regulation of protein kinase A signaling / negative regulation of osteoclast differentiation / Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly / bone resorption ...amylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption / eating behavior / positive regulation of protein kinase A signaling / negative regulation of osteoclast differentiation / Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly / bone resorption / sensory perception of pain / positive regulation of calcium-mediated signaling / osteoclast differentiation / hormone activity / cell-cell signaling / amyloid-beta binding / G alpha (s) signalling events / positive regulation of MAPK cascade / receptor ligand activity / positive regulation of apoptotic process / Amyloid fiber formation / signaling receptor binding / lipid binding / apoptotic process / signal transduction / extracellular space / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | ELECTRON MICROSCOPY / helical reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Gallardo, R.U. / Iadanza, M.G. / Ranson, N.A. / Radford, S.E. | |||||||||
| Funding support |  United Kingdom, 2items United Kingdom, 2items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: Fibril structures of diabetes-related amylin variants reveal a basis for surface-templated assembly. Authors: Rodrigo Gallardo / Matthew G Iadanza / Yong Xu / George R Heath / Richard Foster / Sheena E Radford / Neil A Ranson /  Abstract: Aggregation of the peptide hormone amylin into amyloid deposits is a pathological hallmark of type-2 diabetes (T2D). While no causal link between T2D and amyloid has been established, the S20G ...Aggregation of the peptide hormone amylin into amyloid deposits is a pathological hallmark of type-2 diabetes (T2D). While no causal link between T2D and amyloid has been established, the S20G mutation in amylin is associated with early-onset T2D. Here we report cryo-EM structures of amyloid fibrils of wild-type human amylin and its S20G variant. The wild-type fibril structure, solved to 3.6-Å resolution, contains two protofilaments, each built from S-shaped subunits. S20G fibrils, by contrast, contain two major polymorphs. Their structures, solved at 3.9-Å and 4.0-Å resolution, respectively, share a common two-protofilament core that is distinct from the wild-type structure. Remarkably, one polymorph contains a third subunit with another, distinct, cross-β conformation. The presence of two different backbone conformations within the same fibril may explain the increased aggregation propensity of S20G, and illustrates a potential structural basis for surface-templated fibril assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6zrq.cif.gz 6zrq.cif.gz | 86.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6zrq.ent.gz pdb6zrq.ent.gz | 74.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6zrq.json.gz 6zrq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6zrq_validation.pdf.gz 6zrq_validation.pdf.gz | 706.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6zrq_full_validation.pdf.gz 6zrq_full_validation.pdf.gz | 715.8 KB | Display | |
| Data in XML |  6zrq_validation.xml.gz 6zrq_validation.xml.gz | 20.4 KB | Display | |
| Data in CIF |  6zrq_validation.cif.gz 6zrq_validation.cif.gz | 29.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zr/6zrq https://data.pdbj.org/pub/pdb/validation_reports/zr/6zrq ftp://data.pdbj.org/pub/pdb/validation_reports/zr/6zrq ftp://data.pdbj.org/pub/pdb/validation_reports/zr/6zrq | HTTPS FTP |
-Related structure data
| Related structure data |  11382MC  6zrfC  6zrrC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein/peptide | Mass: 3878.293 Da / Num. of mol.: 12 / Mutation: S20G / Source method: obtained synthetically / Details: TYC = C-terminal amidated Tyr / Source: (synth.)  Homo sapiens (human) / References: UniProt: P10997 Homo sapiens (human) / References: UniProt: P10997Has ligand of interest | N | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method: helical reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | ||||||||||||||||||
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Source (recombinant) | Organism: synthetic construct (others) | ||||||||||||||||||
| Buffer solution | pH: 6.8 | ||||||||||||||||||
| Buffer component | Conc.: 20 mM / Name: ammonium acetate / Formula: NH4CH3CO | ||||||||||||||||||
| Specimen | Conc.: 0.12 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: Synthetically produced, oxidised and C-terminally amidated | ||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Homemade | ||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 85 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 130000 X / Cs: 2.7 mm / C2 aperture diameter: 100 µm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 13 sec. / Electron dose: 54.73 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
| EM imaging optics | Energyfilter name: GIF Quantum LS / Energyfilter slit width: 20 eV |
| Image scans | Movie frames/image: 52 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||
| Helical symmerty | Angular rotation/subunit: 179.05 ° / Axial rise/subunit: 2.41 Å / Axial symmetry: C1 | ||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 64274 | ||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 11901 / Num. of class averages: 1 / Symmetry type: HELICAL |
 Movie
Movie Controller
Controller












 PDBj
PDBj



