[English] 日本語
 Yorodumi
Yorodumi- PDB-6caj: Electron cryo-microscopy of the eukaryotic translation initiation... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6caj | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B from Homo sapiens | |||||||||
 Components Components | (Translation initiation factor eIF-2B subunit ...) x 5 | |||||||||
 Keywords Keywords | TRANSLATION / guanine nucleotide exchange factor / translation initiation / ISRIB-bound / decameric complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationeukaryotic translation initiation factor 2B complex / Recycling of eIF2:GDP / cytoplasmic translational initiation / astrocyte development / guanyl-nucleotide exchange factor complex / astrocyte differentiation / oligodendrocyte development / regulation of translational initiation / positive regulation of translational initiation / response to glucose ...eukaryotic translation initiation factor 2B complex / Recycling of eIF2:GDP / cytoplasmic translational initiation / astrocyte development / guanyl-nucleotide exchange factor complex / astrocyte differentiation / oligodendrocyte development / regulation of translational initiation / positive regulation of translational initiation / response to glucose / ovarian follicle development / myelination / translation initiation factor binding / translation initiation factor activity / guanyl-nucleotide exchange factor activity / response to endoplasmic reticulum stress / central nervous system development / hippocampus development / translational initiation / response to peptide hormone / T cell receptor signaling pathway / regulation of translation / response to heat / positive regulation of apoptotic process / GTP binding / ATP binding / identical protein binding / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Tsai, J.C. / Miller-Vedam, L.E. / Anand, A.A. / Jaishankar, P. / Nguyen, H.C. / Renslo, A.R. / Frost, A. / Walter, P. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Authors: Jordan C Tsai / Lakshmi E Miller-Vedam / Aditya A Anand / Priyadarshini Jaishankar / Henry C Nguyen / Adam R Renslo / Adam Frost / Peter Walter /  Abstract: Regulation by the integrated stress response (ISR) converges on the phosphorylation of translation initiation factor eIF2 in response to a variety of stresses. Phosphorylation converts eIF2 from a ...Regulation by the integrated stress response (ISR) converges on the phosphorylation of translation initiation factor eIF2 in response to a variety of stresses. Phosphorylation converts eIF2 from a substrate to a competitive inhibitor of its dedicated guanine nucleotide exchange factor, eIF2B, thereby inhibiting translation. ISRIB, a drug-like eIF2B activator, reverses the effects of eIF2 phosphorylation, and in rodents it enhances cognition and corrects cognitive deficits after brain injury. To determine its mechanism of action, we solved an atomic-resolution structure of ISRIB bound in a deep cleft within decameric human eIF2B by cryo-electron microscopy. Formation of fully active, decameric eIF2B holoenzyme depended on the assembly of two identical tetrameric subcomplexes, and ISRIB promoted this step by cross-bridging a central symmetry interface. Thus, regulation of eIF2B assembly emerges as a rheostat for eIF2B activity that tunes translation during the ISR and that can be further modulated by ISRIB. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6caj.cif.gz 6caj.cif.gz | 571.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6caj.ent.gz pdb6caj.ent.gz | 441 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6caj.json.gz 6caj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ca/6caj https://data.pdbj.org/pub/pdb/validation_reports/ca/6caj ftp://data.pdbj.org/pub/pdb/validation_reports/ca/6caj ftp://data.pdbj.org/pub/pdb/validation_reports/ca/6caj | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7442MC  7443C  7444C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Translation initiation factor eIF-2B subunit ... , 5 types, 10 molecules BAJIHGCDFE
| #1: Protein | Mass: 80452.586 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: EIF2B5, EIF2BE / Production host: Homo sapiens (human) / Gene: EIF2B5, EIF2BE / Production host:  #2: Protein | Mass: 50304.230 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: EIF2B3 / Production host: Homo sapiens (human) / Gene: EIF2B3 / Production host:  #3: Protein | Mass: 33754.148 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: EIF2B1, EIF2BA / Production host: Homo sapiens (human) / Gene: EIF2B1, EIF2BA / Production host:  #4: Protein | Mass: 41008.578 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: EIF2B2, EIF2BB / Production host: Homo sapiens (human) / Gene: EIF2B2, EIF2BB / Production host:  #5: Protein | Mass: 57640.168 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: EIF2B4, EIF2BD / Production host: Homo sapiens (human) / Gene: EIF2B4, EIF2BD / Production host:  |
|---|
-Non-polymers , 1 types, 1 molecules 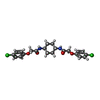
| #6: Chemical | ChemComp-C7B / |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Translation initiation factor eIF-2B decamer / Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 1.63 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 202125 | ||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C2 (2 fold cyclic) | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.8 Å / Resolution method: FSC 0.5 CUT-OFF / Num. of particles: 202125 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL |
 Movie
Movie Controller
Controller









 PDBj
PDBj






