+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 6usv | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | |||||||||
 要素 要素 | (Glutamate receptor ionotropic, NMDA ...) x 2 | |||||||||
 キーワード キーワード | METAL TRANSPORT / NMDARs / LBD / Ion channels | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報neurotransmitter receptor transport, plasma membrane to endosome / regulation of response to alcohol / response to ammonium ion / receptor recycling / response to environmental enrichment / directional locomotion / positive regulation of Schwann cell migration / pons maturation / regulation of cell communication / EPHB-mediated forward signaling ...neurotransmitter receptor transport, plasma membrane to endosome / regulation of response to alcohol / response to ammonium ion / receptor recycling / response to environmental enrichment / directional locomotion / positive regulation of Schwann cell migration / pons maturation / regulation of cell communication / EPHB-mediated forward signaling / Assembly and cell surface presentation of NMDA receptors / response to hydrogen sulfide / auditory behavior / olfactory learning / conditioned taste aversion / dendritic branch / regulation of respiratory gaseous exchange / regulation of ARF protein signal transduction / response to other organism / protein localization to postsynaptic membrane / cellular response to magnesium ion / serotonin metabolic process / transmitter-gated monoatomic ion channel activity / positive regulation of inhibitory postsynaptic potential / response to methylmercury / suckling behavior / response to manganese ion / response to glycine / response to carbohydrate / propylene metabolic process / sleep / dendritic spine organization / cellular response to dsRNA / locomotion / regulation of NMDA receptor activity / cellular response to lipid / RAF/MAP kinase cascade / regulation of monoatomic cation transmembrane transport / NMDA glutamate receptor activity / Synaptic adhesion-like molecules / response to glycoside / voltage-gated monoatomic cation channel activity / NMDA selective glutamate receptor complex / glutamate binding / neurotransmitter receptor complex / ligand-gated sodium channel activity / response to morphine / glutamate receptor signaling pathway / regulation of axonogenesis / calcium ion transmembrane import into cytosol / neuromuscular process / regulation of dendrite morphogenesis / protein heterotetramerization / male mating behavior / regulation of synapse assembly / spinal cord development / response to amine / glycine binding / cellular response to zinc ion / startle response / positive regulation of reactive oxygen species biosynthetic process / dopamine metabolic process / parallel fiber to Purkinje cell synapse / response to lithium ion / monoatomic cation transmembrane transport / positive regulation of calcium ion transport into cytosol / cellular response to glycine / regulation of postsynaptic membrane potential / response to light stimulus / modulation of excitatory postsynaptic potential / action potential / associative learning / conditioned place preference / positive regulation of dendritic spine maintenance / monoatomic ion channel complex / regulation of neuronal synaptic plasticity / monoatomic cation transport / social behavior / positive regulation of protein targeting to membrane / glutamate receptor binding / Unblocking of NMDA receptors, glutamate binding and activation / long-term memory / synaptic cleft / neuron development / phosphatase binding / prepulse inhibition / positive regulation of synaptic transmission, glutamatergic / multicellular organismal response to stress / postsynaptic density, intracellular component / monoatomic cation channel activity / response to fungicide / calcium ion homeostasis / glutamate-gated receptor activity / regulation of neuron apoptotic process / cell adhesion molecule binding / cellular response to manganese ion / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / neurogenesis / dendrite membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.304 Å 分子置換 / 解像度: 2.304 Å | |||||||||
 データ登録者 データ登録者 | Romero-Hernandez, A. / Tajima, N. / Chou, T. / Furukawa, h. | |||||||||
| 資金援助 |  米国, 2件 米国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Cell / 年: 2020 ジャーナル: Cell / 年: 2020タイトル: Structural Basis of Functional Transitions in Mammalian NMDA Receptors. 著者: Tsung-Han Chou / Nami Tajima / Annabel Romero-Hernandez / Hiro Furukawa /  要旨: Excitatory neurotransmission meditated by glutamate receptors including N-methyl-D-aspartate receptors (NMDARs) is pivotal to brain development and function. NMDARs are heterotetramers composed of ...Excitatory neurotransmission meditated by glutamate receptors including N-methyl-D-aspartate receptors (NMDARs) is pivotal to brain development and function. NMDARs are heterotetramers composed of GluN1 and GluN2 subunits, which bind glycine and glutamate, respectively, to activate their ion channels. Despite importance in brain physiology, the precise mechanisms by which activation and inhibition occur via subunit-specific binding of agonists and antagonists remain largely unknown. Here, we show the detailed patterns of conformational changes and inter-subunit and -domain reorientation leading to agonist-gating and subunit-dependent competitive inhibition by providing multiple structures in distinct ligand states at 4 Å or better. The structures reveal that activation and competitive inhibition by both GluN1 and GluN2 antagonists occur by controlling the tension of the linker between the ligand-binding domain and the transmembrane ion channel of the GluN2 subunit. Our results provide detailed mechanistic insights into NMDAR pharmacology, activation, and inhibition, which are fundamental to the brain physiology. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  6usv.cif.gz 6usv.cif.gz | 128.6 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb6usv.ent.gz pdb6usv.ent.gz | 96.1 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  6usv.json.gz 6usv.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/us/6usv https://data.pdbj.org/pub/pdb/validation_reports/us/6usv ftp://data.pdbj.org/pub/pdb/validation_reports/us/6usv ftp://data.pdbj.org/pub/pdb/validation_reports/us/6usv | HTTPS FTP |
|---|
-関連構造データ
| 関連構造データ |  6usuC  6whrC  6whsC  6whtC  6whuC  6whvC  6whwC  6whxC  6whyC  6wi0C  6wi1C  4nf8S S: 精密化の開始モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| 単位格子 |
|
- 要素
要素
-Glutamate receptor ionotropic, NMDA ... , 2種, 2分子 AB
| #1: タンパク質 | 分子量: 33340.031 Da / 分子数: 1 / 断片: UNP residues 415-565, 684-821 / 由来タイプ: 組換発現 / 由来: (組換発現)   |
|---|---|
| #2: タンパク質 | 分子量: 31785.299 Da / 分子数: 1 / 断片: UNP residues 402-539, 661-802 / 由来タイプ: 組換発現 / 由来: (組換発現)   |
-非ポリマー , 4種, 45分子 
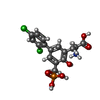





| #3: 化合物 | ChemComp-GLY / |
|---|---|
| #4: 化合物 | ChemComp-QGP / ( |
| #5: 化合物 | ChemComp-GOL / |
| #6: 水 | ChemComp-HOH / |
-詳細
| 研究の焦点であるリガンドがあるか | Y |
|---|---|
| Has protein modification | Y |
-実験情報
-実験
| 実験 | 手法:  X線回折 / 使用した結晶の数: 1 X線回折 / 使用した結晶の数: 1 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 2.36 Å3/Da / 溶媒含有率: 47.82 % |
|---|---|
| 結晶化 | 温度: 291 K / 手法: 蒸発脱水法 / pH: 7 詳細: 0.2 M HEPES, pH 7.0, 60-90 mM sodium chloride, 15-20% PEG2000 MME |
-データ収集
| 回折 | 平均測定温度: 100 K / Serial crystal experiment: N |
|---|---|
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  APS APS  / ビームライン: 23-ID-B / 波長: 0.97 Å / ビームライン: 23-ID-B / 波長: 0.97 Å |
| 検出器 | タイプ: DECTRIS EIGER X 16M / 検出器: PIXEL / 日付: 2013年11月11日 |
| 放射 | モノクロメーター: Double crystal cryo-cooled Si(111) プロトコル: SINGLE WAVELENGTH / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray |
| 放射波長 | 波長: 0.97 Å / 相対比: 1 |
| 反射 | 解像度: 2.3→49.5 Å / Num. obs: 26575 / % possible obs: 95 % / 冗長度: 4.8 % / Biso Wilson estimate: 44.07 Å2 / Rpim(I) all: 0.042 / Rsym value: 0.082 / Net I/σ(I): 9.5 |
| 反射 シェル | 解像度: 2.3→2.34 Å / Num. unique obs: 1276 / CC1/2: 0.872 / Rpim(I) all: 0.579 |
- 解析
解析
| ソフトウェア |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 構造決定の手法:  分子置換 分子置換開始モデル: PDB entry 4NF8 解像度: 2.304→49.497 Å / SU ML: 0.35 / 交差検証法: THROUGHOUT / σ(F): 1.34 / 位相誤差: 28.37
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | 減衰半径: 0.9 Å / VDWプローブ半径: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | Biso max: 86.96 Å2 / Biso mean: 45.8 Å2 / Biso min: 22.81 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: final / 解像度: 2.304→49.497 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 拘束条件 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS精密化 シェル | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0
|
 ムービー
ムービー コントローラー
コントローラー























 PDBj
PDBj







