[English] 日本語
 Yorodumi
Yorodumi- EMDB-6478: Single-particle cryo-electron microscopy reconstruction of the eu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6478 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-particle cryo-electron microscopy reconstruction of the eukaryotic large ribosomal subunit | |||||||||
 Map data Map data | Single-particle cryo-EM reconstruction of the eukaryotic large 60S ribosomal subunit | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / translation | |||||||||
| Function / homology |  Function and homology information Function and homology informationSRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / Formation of a pool of free 40S subunits / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / L13a-mediated translational silencing of Ceruloplasmin expression / ribosomal large subunit assembly / cytosolic large ribosomal subunit / cytoplasmic translation / structural constituent of ribosome ...SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / Formation of a pool of free 40S subunits / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / L13a-mediated translational silencing of Ceruloplasmin expression / ribosomal large subunit assembly / cytosolic large ribosomal subunit / cytoplasmic translation / structural constituent of ribosome / RNA binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Passos DO / Lyumkis D | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2015 Journal: J Struct Biol / Year: 2015Title: Single-particle cryoEM analysis at near-atomic resolution from several thousand asymmetric subunits. Authors: Dario Oliveira Passos / Dmitry Lyumkis /  Abstract: A single-particle cryoEM reconstruction of the large ribosomal subunit from Saccharomyces cerevisiae was obtained from a dataset of ∼75,000 particles. The gold-standard and frequency-limited ...A single-particle cryoEM reconstruction of the large ribosomal subunit from Saccharomyces cerevisiae was obtained from a dataset of ∼75,000 particles. The gold-standard and frequency-limited approaches to single-particle refinement were each independently used to determine orientation parameters for the final reconstruction. Both approaches showed similar resolution curves and nominal resolution values for the 60S dataset, estimated at 2.9 Å. The amount of over-fitting present during frequency-limited refinement was quantitatively analyzed using the high-resolution phase-randomization test, and the results showed no apparent over-fitting. The number of asymmetric subunits required to reach specific resolutions was subsequently analyzed by refining subsets of the data in an ab initio manner. With our data collection and processing strategies, sub-nanometer resolution was obtained with ∼200 asymmetric subunits (or, equivalently for the ribosomal subunit, particles). Resolutions of 5.6 Å, 4.5 Å, and 3.8 Å were reached with ∼1000, ∼1600, and ∼5000 asymmetric subunits, respectively. At these resolutions, one would expect to detect alpha-helical pitch, separation of beta-strands, and separation of Cα atoms, respectively. Using this map, together with strategies for ab initio model building and model refinement, we built a region of the ribosomal protein eL6, which was missing in previous models of the yeast ribosome. The relevance for more routine high-resolution structure determination is discussed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6478.map.gz emd_6478.map.gz | 104.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6478-v30.xml emd-6478-v30.xml emd-6478.xml emd-6478.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6478.gif 400_6478.gif 80_6478.gif 80_6478.gif | 80.2 KB 4.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6478 http://ftp.pdbj.org/pub/emdb/structures/EMD-6478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6478 | HTTPS FTP |
-Related structure data
| Related structure data |  3jbsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6478.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6478.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM reconstruction of the eukaryotic large 60S ribosomal subunit | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
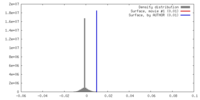
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae large 60S ribosomal subunit
| Entire | Name: Saccharomyces cerevisiae large 60S ribosomal subunit |
|---|---|
| Components |
|
-Supramolecule #1000: Saccharomyces cerevisiae large 60S ribosomal subunit
| Supramolecule | Name: Saccharomyces cerevisiae large 60S ribosomal subunit / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 2.5 MDa |
-Supramolecule #1: 60S large ribosomal subunit
| Supramolecule | Name: 60S large ribosomal subunit / type: complex / ID: 1 / Recombinant expression: No / Database: NCBI / Ribosome-details: ribosome-eukaryote: LSU 60S |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 Details: 50 mM HEPES-KOH, 100 mM KOAc, 5 mM MgOAc, 1 mM EDTA, 2 mM DTT |
|---|---|
| Grid | Details: 400 mesh, 1.2x1.3 micron C-flat grids, plasma-treated for 5 seconds |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 77 K / Instrument: HOMEMADE PLUNGER Method: A 3 uL sample was applied onto a freshly plasma-treated (6 seconds, Gatan Solarus plasma cleaner) holey carbon C-flat grid (Protochips, Inc.), allowing the sample to adsorb for 30 seconds. ...Method: A 3 uL sample was applied onto a freshly plasma-treated (6 seconds, Gatan Solarus plasma cleaner) holey carbon C-flat grid (Protochips, Inc.), allowing the sample to adsorb for 30 seconds. The sample was then plunge-frozen in liquid ethane using a manual cryo-plunger in ambient atmosphere at 4 degrees C. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected within the Leginon software. |
| Details | super-resolution imaging |
| Date | Sep 2, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 1833 / Average electron dose: 25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 38167 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: OTHER / Software - Name: Frealign / Details: B-factor of -50 applied to final map / Number images used: 75653 |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)