[English] 日本語
 Yorodumi
Yorodumi- EMDB-5995: Structure of beta-galactosidase at 3.2-A resolution obtained by c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5995 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | |||||||||
 Map data Map data | Reconstruction of beta-galactosidase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | atomic resolution cryo-electron microscopy / single-particle EM / direct electron detectors / 3D reconstruction / frame alignment / CTF determination / structure refinement / radiation damage / protein complexes / enzyme active site structure | |||||||||
| Function / homology |  Function and homology information Function and homology informationalkali metal ion binding / lactose catabolic process / beta-galactosidase complex / beta-galactosidase / beta-galactosidase activity / carbohydrate binding / magnesium ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Bartesaghi A / Matthies D / Banerjee S / Merk A / Subramaniam S | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2014 Journal: Proc Natl Acad Sci U S A / Year: 2014Title: Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Authors: Alberto Bartesaghi / Doreen Matthies / Soojay Banerjee / Alan Merk / Sriram Subramaniam /  Abstract: We report the solution structure of Escherichia coli β-galactosidase (∼465 kDa), solved at ∼3.2-Å resolution by using single-particle cryo-electron microscopy (cryo-EM). Densities for most side ...We report the solution structure of Escherichia coli β-galactosidase (∼465 kDa), solved at ∼3.2-Å resolution by using single-particle cryo-electron microscopy (cryo-EM). Densities for most side chains, including those of residues in the active site, and a catalytic Mg(2+) ion can be discerned in the map obtained by cryo-EM. The atomic model derived from our cryo-EM analysis closely matches the 1.7-Å crystal structure with a global rmsd of ∼0.66 Å. There are significant local differences throughout the protein, with clear evidence for conformational changes resulting from contact zones in the crystal lattice. Inspection of the map reveals that although densities for residues with positively charged and neutral side chains are well resolved, systematically weaker densities are observed for residues with negatively charged side chains. We show that the weaker densities for negatively charged residues arise from their greater sensitivity to radiation damage from electron irradiation as determined by comparison of density maps obtained by using electron doses ranging from 10 to 30 e(-)/Å(2). In summary, we establish that it is feasible to use cryo-EM to determine near-atomic resolution structures of protein complexes (<500 kDa) with low symmetry, and that the residue-specific radiation damage that occurs with increasing electron dose can be monitored by using dose fractionation tools available with direct electron detector technology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5995.map.gz emd_5995.map.gz | 136.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5995-v30.xml emd-5995-v30.xml emd-5995.xml emd-5995.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_5995_fsc.xml emd_5995_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  400_5995.gif 400_5995.gif 80_5995.gif 80_5995.gif | 82.5 KB 4.5 KB | ||
| Others |  emd_5995_additional_1.map.gz emd_5995_additional_1.map.gz emd_5995_half_map_1.map.gz emd_5995_half_map_1.map.gz emd_5995_half_map_2.map.gz emd_5995_half_map_2.map.gz | 59.7 MB 61.2 MB 61.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5995 http://ftp.pdbj.org/pub/emdb/structures/EMD-5995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5995 | HTTPS FTP |
-Related structure data
| Related structure data |  3j7hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10012 (Title: Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy (frame-averaged micrographs) EMPIAR-10012 (Title: Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy (frame-averaged micrographs)Data size: 108.0 Data #1: Beta Galactosidase micrographs [micrographs - single frame])  EMPIAR-10013 (Title: Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy EMPIAR-10013 (Title: Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopyData size: 442.5 Data #1: Full set of multi-frame micrographs [micrographs - multiframe] Data #2: Sample set of multi-frame micrographs [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5995.map.gz / Format: CCP4 / Size: 146.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5995.map.gz / Format: CCP4 / Size: 146.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of beta-galactosidase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.6375 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 5995 additional 1.map
| File | emd_5995_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
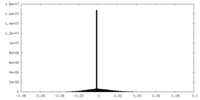
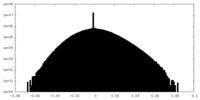
| Density Histograms |
-Supplemental map: emd 5995 half map 1.map
| File | emd_5995_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 5995 half map 2.map
| File | emd_5995_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia coli beta-galactosidase
| Entire | Name: Escherichia coli beta-galactosidase |
|---|---|
| Components |
|
-Supramolecule #1000: Escherichia coli beta-galactosidase
| Supramolecule | Name: Escherichia coli beta-galactosidase / type: sample / ID: 1000 / Details: The sample was monodisperse. / Oligomeric state: tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 465 KDa Method: ProtParam tool: Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A., Protein Identification and Analysis Tools on the ExPASy Server, (in) John M. Walker ...Method: ProtParam tool: Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A., Protein Identification and Analysis Tools on the ExPASy Server, (in) John M. Walker (ed): The Proteomics Protocols Handbook, Humana Press (2005), pp. 571-607 |
-Macromolecule #1: beta-galactosidase
| Macromolecule | Name: beta-galactosidase / type: protein_or_peptide / ID: 1 / Name.synonym: beta-gal, b-gal / Number of copies: 1 / Oligomeric state: tetramer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 465 KDa |
| Sequence | UniProtKB: Beta-galactosidase / GO: beta-galactosidase activity |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.3 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 25 mM Tris, pH 8.0, 50 mM NaCl, 2 mM MgCl2, 0.5 mM TCEP |
| Grid | Details: 200 mesh Quantifoil R2/2 grids, plasma cleaned |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 90.15 K / Instrument: LEICA EM GP / Method: Blot for 2 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.6 K / Max: 79.8 K / Average: 79.7 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 105,000 times magnification. Legacy - Electron beam tilt params: 5 |
| Specialist optics | Energy filter - Name: Gatan, Inc. / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Details | Parallel beam illumination |
| Date | Oct 31, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 509 / Average electron dose: 45 e/Å2 Details: Every image is the average of 38 frames recorded by the direct electron detector. The complete set of electron micrographs used to obtain the density map presented here is available through ...Details: Every image is the average of 38 frames recorded by the direct electron detector. The complete set of electron micrographs used to obtain the density map presented here is available through the Electron Microscopy Pilot Image ARchive (EMPIAR). |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)