+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7770 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Atomic resolution cryo-EM structure of beta-galactosidase | |||||||||
 Map data Map data | Escherichia coli beta-galactosidase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | drift correction / radiation damage / drug discovery / precision medicine / computer-aided drug discovery / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationalkali metal ion binding / lactose catabolic process / beta-galactosidase complex / beta-galactosidase / beta-galactosidase activity / carbohydrate binding / magnesium ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
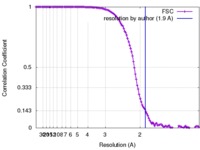
| Method | single particle reconstruction / cryo EM / Resolution: 1.9 Å | |||||||||
 Authors Authors | Subramaniam S / Bartesaghi A / Banerjee S / Zhu X | |||||||||
 Citation Citation |  Journal: Structure / Year: 2018 Journal: Structure / Year: 2018Title: Atomic Resolution Cryo-EM Structure of β-Galactosidase. Authors: Alberto Bartesaghi / Cecilia Aguerrebere / Veronica Falconieri / Soojay Banerjee / Lesley A Earl / Xing Zhu / Nikolaus Grigorieff / Jacqueline L S Milne / Guillermo Sapiro / Xiongwu Wu / Sriram Subramaniam /  Abstract: The advent of direct electron detectors has enabled the routine use of single-particle cryo-electron microscopy (EM) approaches to determine structures of a variety of protein complexes at near- ...The advent of direct electron detectors has enabled the routine use of single-particle cryo-electron microscopy (EM) approaches to determine structures of a variety of protein complexes at near-atomic resolution. Here, we report the development of methods to account for local variations in defocus and beam-induced drift, and the implementation of a data-driven dose compensation scheme that significantly improves the extraction of high-resolution information recorded during exposure of the specimen to the electron beam. These advances enable determination of a cryo-EM density map for β-galactosidase bound to the inhibitor phenylethyl β-D-thiogalactopyranoside where the ordered regions are resolved at a level of detail seen in X-ray maps at ∼ 1.5 Å resolution. Using this density map in conjunction with constrained molecular dynamics simulations provides a measure of the local flexibility of the non-covalently bound inhibitor and offers further opportunities for structure-guided inhibitor design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7770.map.gz emd_7770.map.gz | 70.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7770-v30.xml emd-7770-v30.xml emd-7770.xml emd-7770.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7770_fsc.xml emd_7770_fsc.xml | 13.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_7770.png emd_7770.png | 197.7 KB | ||
| Masks |  emd_7770_msk_1.map emd_7770_msk_1.map | 147.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-7770.cif.gz emd-7770.cif.gz | 6.5 KB | ||
| Others |  emd_7770_additional.map.gz emd_7770_additional.map.gz emd_7770_half_map_1.map.gz emd_7770_half_map_1.map.gz emd_7770_half_map_2.map.gz emd_7770_half_map_2.map.gz | 53.3 MB 71 MB 71 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7770 http://ftp.pdbj.org/pub/emdb/structures/EMD-7770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7770 | HTTPS FTP |
-Related structure data
| Related structure data |  6cvmMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7770.map.gz / Format: CCP4 / Size: 147.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7770.map.gz / Format: CCP4 / Size: 147.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Escherichia coli beta-galactosidase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.637 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
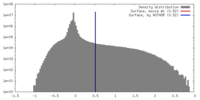
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7770_msk_1.map emd_7770_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Escherichia coli beta-galactosidase, sharpened map
| File | emd_7770_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Escherichia coli beta-galactosidase, sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Escherichia coli beta-galactosidase, half map #1
| File | emd_7770_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Escherichia coli beta-galactosidase, half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Escherichia coli beta-galactosidase, half map #2
| File | emd_7770_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Escherichia coli beta-galactosidase, half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia coli beta-galactosidase bound to phenylethyl beta-D-t...
| Entire | Name: Escherichia coli beta-galactosidase bound to phenylethyl beta-D-thiogalactopyranoside (PETG) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia coli beta-galactosidase bound to phenylethyl beta-D-t...
| Supramolecule | Name: Escherichia coli beta-galactosidase bound to phenylethyl beta-D-thiogalactopyranoside (PETG) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 465 KDa |
-Macromolecule #1: Beta-galactosidase
| Macromolecule | Name: Beta-galactosidase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: beta-galactosidase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 116.181031 KDa |
| Sequence | String: MITDSLAVVL QRRDWENPGV TQLNRLAAHP PFASWRNSEE ARTDRPSQQL RSLNGEWRFA WFPAPEAVPE SWLECDLPEA DTVVVPSNW QMHGYDAPIY TNVTYPITVN PPFVPTENPT GCYSLTFNVD ESWLQEGQTR IIFDGVNSAF HLWCNGRWVG Y GQDSRLPS ...String: MITDSLAVVL QRRDWENPGV TQLNRLAAHP PFASWRNSEE ARTDRPSQQL RSLNGEWRFA WFPAPEAVPE SWLECDLPEA DTVVVPSNW QMHGYDAPIY TNVTYPITVN PPFVPTENPT GCYSLTFNVD ESWLQEGQTR IIFDGVNSAF HLWCNGRWVG Y GQDSRLPS EFDLSAFLRA GENRLAVMVL RWSDGSYLED QDMWRMSGIF RDVSLLHKPT TQISDFHVAT RFNDDFSRAV LE AEVQMCG ELRDYLRVTV SLWQGETQVA SGTAPFGGEI IDERGGYADR VTLRLNVENP KLWSAEIPNL YRAVVELHTA DGT LIEAEA CDVGFRVVRI ENGLLLLNGK PLLIRGVNRH EHHPLHGQVM DEQTMVQDIL LMKQNNFNAV RCSHYPNHPL WYTL CDRYG LYVVDEANIE THGMVPMNRL TDDPRWLPAM SERVTRMVQR DRNHPSVIIW SLGNESGHGA NHDALYRWIK SVDPS RPVQ YEGGGADTTA TDIICPMYAR VDEDQPFPAV PKWSIKKWLS LPGETRPLIL CEYAHAMGNS LGGFAKYWQA FRQYPR LQG GFVWDWVDQS LIKYDENGNP WSAYGGDFGD TPNDRQFCMN GLVFADRTPH PALTEAKHQQ QFFQFRLSGQ TIEVTSE YL FRHSDNELLH WMVALDGKPL ASGEVPLDVA PQGKQLIELP ELPQPESAGQ LWLTVRVVQP NATAWSEAGH ISAWQQWR L AENLSVTLPA ASHAIPHLTT SEMDFCIELG NKRWQFNRQS GFLSQMWIGD KKQLLTPLRD QFTRAPLDND IGVSEATRI DPNAWVERWK AAGHYQAEAA LLQCTADTLA DAVLITTAHA WQHQGKTLFI SRKTYRIDGS GQMAITVDVV VASDTPHPAR IGLNCQLAQ VAERVNWLGL GPQENYPDRL TAACFDRWDL PLSDMYTPYV FPSENGLRCG TRELNYGPHQ WRGDFQFNIS R YSQQQLME TSHRHLLHAE EGTWLNIDGF HMGIGGDDSW SPSVSAEFQL SAGRYHYQLV WCQ UniProtKB: Beta-galactosidase |
-Macromolecule #2: 2-phenylethyl 1-thio-beta-D-galactopyranoside
| Macromolecule | Name: 2-phenylethyl 1-thio-beta-D-galactopyranoside / type: ligand / ID: 2 / Number of copies: 4 / Formula: PTQ |
|---|---|
| Molecular weight | Theoretical: 300.371 Da |
| Chemical component information |  ChemComp-PTQ: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 8 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 8 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 4194 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.3 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 25 mM Tris, pH 8.0, 50 mM NaCl, 2 mM MgCl2, 1.0 mM TCEP |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Details: plasma cleaned |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 90.15 K / Instrument: LEICA EM GP / Details: Blot for 2 seconds before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.6 K / Max: 79.8 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | Parallel beam illumination |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-38 / Number real images: 1539 / Average exposure time: 7.6 sec. / Average electron dose: 45.0 e/Å2 / Details: Raw micrographs are available from EMPIAR-10061. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.0 µm / Calibrated magnification: 215000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: 0.6 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6cvm: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)