[English] 日本語
 Yorodumi
Yorodumi- EMDB-4316: Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Boun... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4316 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Protein translocon of the endoplasmic reticulum / PROTEIN TRANSPORT | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationoligosaccharyltransferase complex A / oligosaccharyltransferase complex B / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / membrane docking / endoplasmic reticulum Sec complex / pronephric nephron development / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / cotranslational protein targeting to membrane ...oligosaccharyltransferase complex A / oligosaccharyltransferase complex B / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / membrane docking / endoplasmic reticulum Sec complex / pronephric nephron development / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / cotranslational protein targeting to membrane / Ssh1 translocon complex / Sec61 translocon complex / protein targeting to ER / protein insertion into ER membrane / protein-transporting ATPase activity / protein N-linked glycosylation via asparagine / : / SRP-dependent cotranslational protein targeting to membrane, translocation / signal sequence binding / post-translational protein targeting to membrane, translocation / ribosomal subunit / : / ubiquitin ligase inhibitor activity / positive regulation of signal transduction by p53 class mediator / protein transmembrane transporter activity / rough endoplasmic reticulum / MDM2/MDM4 family protein binding / cytosolic ribosome / post-translational protein modification / guanyl-nucleotide exchange factor activity / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal large subunit biogenesis / phospholipid binding / ribosome biogenesis / large ribosomal subunit / regulation of translation / ribosome binding / 5S rRNA binding / ribosomal large subunit assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / synapse / endoplasmic reticulum membrane / nucleolus / endoplasmic reticulum / RNA binding / zinc ion binding / metal ion binding / nucleus / membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |   | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | ||||||||||||||||||
 Authors Authors | Braunger K / Becker T | ||||||||||||||||||
| Funding support |  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Authors: Katharina Braunger / Stefan Pfeffer / Shiteshu Shrimal / Reid Gilmore / Otto Berninghausen / Elisabet C Mandon / Thomas Becker / Friedrich Förster / Roland Beckmann /    Abstract: Protein synthesis, transport, and N-glycosylation are coupled at the mammalian endoplasmic reticulum by complex formation of a ribosome, the Sec61 protein-conducting channel, and ...Protein synthesis, transport, and N-glycosylation are coupled at the mammalian endoplasmic reticulum by complex formation of a ribosome, the Sec61 protein-conducting channel, and oligosaccharyltransferase (OST). Here we used different cryo-electron microscopy approaches to determine structures of native and solubilized ribosome-Sec61-OST complexes. A molecular model for the catalytic OST subunit STT3A (staurosporine and temperature sensitive 3A) revealed how it is integrated into the OST and how STT3-paralog specificity for translocon-associated OST is achieved. The OST subunit DC2 was placed at the interface between Sec61 and STT3A, where it acts as a versatile module for recruitment of STT3A-containing OST to the ribosome-Sec61 complex. This detailed structural view on the molecular architecture of the cotranslational machinery for N-glycosylation provides the basis for a mechanistic understanding of glycoprotein biogenesis at the endoplasmic reticulum. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4316.map.gz emd_4316.map.gz | 442.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4316-v30.xml emd-4316-v30.xml emd-4316.xml emd-4316.xml | 107.5 KB 107.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4316.png emd_4316.png | 76.1 KB | ||
| Masks |  emd_4316_msk_1.map emd_4316_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4316.cif.gz emd-4316.cif.gz | 17.7 KB | ||
| Others |  emd_4316_additional.map.gz emd_4316_additional.map.gz | 445.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4316 http://ftp.pdbj.org/pub/emdb/structures/EMD-4316 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4316 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4316 | HTTPS FTP |
-Validation report
| Summary document |  emd_4316_validation.pdf.gz emd_4316_validation.pdf.gz | 595.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4316_full_validation.pdf.gz emd_4316_full_validation.pdf.gz | 594.7 KB | Display | |
| Data in XML |  emd_4316_validation.xml.gz emd_4316_validation.xml.gz | 7.7 KB | Display | |
| Data in CIF |  emd_4316_validation.cif.gz emd_4316_validation.cif.gz | 9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4316 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4316 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4316 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4316 | HTTPS FTP |
-Related structure data
| Related structure data |  6ftiMC  4306C  4307C  4308C 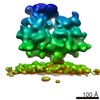 4309C 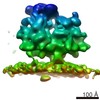 4310C  4311C 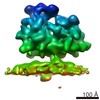 4312C  4313C  4314C  4315C  4317C  6ftgC  6ftjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4316.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4316.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1924 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4316_msk_1.map emd_4316_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_4316_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Mammalian Oligosaccharyltransferase in complex with Sec61 and the...
+Supramolecule #1: Mammalian Oligosaccharyltransferase in complex with Sec61 and the...
+Supramolecule #2: 60S Ribosomal Subunit
+Supramolecule #3: Sec61 protein conducting channel
+Supramolecule #4: Oligosaccharyltransferase
+Macromolecule #1: uL2
+Macromolecule #2: uL3
+Macromolecule #3: Ribosomal protein L4
+Macromolecule #4: 60S ribosomal protein L5
+Macromolecule #5: 60S ribosomal protein L6
+Macromolecule #6: uL30
+Macromolecule #7: eL8
+Macromolecule #8: uL6
+Macromolecule #9: Ribosomal protein L10 (Predicted)
+Macromolecule #10: Ribosomal protein L11
+Macromolecule #11: eL13
+Macromolecule #12: Ribosomal protein L14
+Macromolecule #13: Ribosomal protein L15
+Macromolecule #14: uL13
+Macromolecule #15: uL22
+Macromolecule #16: uL14
+Macromolecule #17: eL19
+Macromolecule #18: eL20
+Macromolecule #19: eL21
+Macromolecule #20: eL22
+Macromolecule #21: uL14
+Macromolecule #22: Ribosomal protein L24
+Macromolecule #23: uL23
+Macromolecule #24: Ribosomal protein L26
+Macromolecule #25: 60S ribosomal protein L27
+Macromolecule #26: uL15
+Macromolecule #27: 60S ribosomal protein L29
+Macromolecule #28: eL30
+Macromolecule #29: eL31
+Macromolecule #30: eL32
+Macromolecule #31: eL33
+Macromolecule #32: eL34
+Macromolecule #33: uL29
+Macromolecule #34: 60S ribosomal protein L36
+Macromolecule #35: Ribosomal protein L37
+Macromolecule #36: eL38
+Macromolecule #37: eL39
+Macromolecule #38: eL40
+Macromolecule #39: 60s ribosomal protein l41
+Macromolecule #40: eL42
+Macromolecule #41: Ribosomal protein L37a
+Macromolecule #42: eL28
+Macromolecule #43: 60S acidic ribosomal protein P0
+Macromolecule #44: Ribosomal protein L12
+Macromolecule #49: Protein transport protein Sec61 subunit alpha isoform 1
+Macromolecule #50: Protein transport protein Sec61 subunit gamma
+Macromolecule #51: Protein transport protein Sec61 subunit beta
+Macromolecule #52: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #53: TMEM258
+Macromolecule #54: Oligosaccharyltransferase complex subunit OSTC
+Macromolecule #55: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #56: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #57: DAD1
+Macromolecule #58: OST48
+Macromolecule #59: RPN1
+Macromolecule #60: Unidentified TM
+Macromolecule #45: p-Site tRNA
+Macromolecule #46: 28S rRNA
+Macromolecule #47: 5S ribosomal RNA
+Macromolecule #48: 5.8S ribosomal RNA
+Macromolecule #62: MAGNESIUM ION
+Macromolecule #63: ZINC ION
+Macromolecule #64: [(2~{S},3~{R},4~{R},5~{S},6~{R})-3-acetamido-6-(hydroxymethyl)-4,...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 28.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)