[English] 日本語
 Yorodumi
Yorodumi- PDB-3s04: Crystal structure of Escherichia coli type I signal peptidase in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3s04 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Escherichia coli type I signal peptidase in complex with an Arylomycin Lipoglycopeptide Antibiotic | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/ANTIBIOTIC / mostly-beta fold / Membrane bound / serine protease / Secreted preproteins / Cytoplasmic membrane / HYDROLASE-ANTIBIOTIC complex / signal peptidase / leader peptidase / signal peptide / leader peptide / serine-lysine dyad | ||||||
| Function / homology |  Function and homology information Function and homology informationsignal peptidase I / : / protein processing / peptidase activity / endopeptidase activity / serine-type endopeptidase activity / proteolysis / plasma membrane Similarity search - Function | ||||||
| Biological species |   Streptomyces sp. (bacteria) Streptomyces sp. (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.44 Å MOLECULAR REPLACEMENT / Resolution: 2.44 Å | ||||||
 Authors Authors | Paetzel, M. / Luo, C. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2011 Journal: J.Am.Chem.Soc. / Year: 2011Title: Synthesis and characterization of the arylomycin lipoglycopeptide antibiotics and the crystallographic analysis of their complex with signal peptidase. Authors: Liu, J. / Luo, C. / Smith, P.A. / Chin, J.K. / Page, M.G. / Paetzel, M. / Romesberg, F.E. | ||||||
| History |
| ||||||
| Remark 300 | THE BIOLOGICAL ASSEMBLY IS A HETERODIMER OF ONE SIGNAL PEPTIDASE I MOLECULE AND ONE ...THE BIOLOGICAL ASSEMBLY IS A HETERODIMER OF ONE SIGNAL PEPTIDASE I MOLECULE AND ONE GLYCOLIPOPEPTIDE INHIBITOR (GLYCO-ARYLOMYCIN) MOLECULE. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3s04.cif.gz 3s04.cif.gz | 197.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3s04.ent.gz pdb3s04.ent.gz | 158.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3s04.json.gz 3s04.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3s04_validation.pdf.gz 3s04_validation.pdf.gz | 480.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3s04_full_validation.pdf.gz 3s04_full_validation.pdf.gz | 482.6 KB | Display | |
| Data in XML |  3s04_validation.xml.gz 3s04_validation.xml.gz | 18.2 KB | Display | |
| Data in CIF |  3s04_validation.cif.gz 3s04_validation.cif.gz | 24.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s0/3s04 https://data.pdbj.org/pub/pdb/validation_reports/s0/3s04 ftp://data.pdbj.org/pub/pdb/validation_reports/s0/3s04 ftp://data.pdbj.org/pub/pdb/validation_reports/s0/3s04 | HTTPS FTP |
-Related structure data
| Related structure data |  1t7dS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 28079.814 Da / Num. of mol.: 2 / Fragment: Periplasmic domain, UNP residues 76-323 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Protein/peptide | 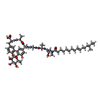  Type: Lipoglycopeptide / Class: Antibiotic / Mass: 660.673 Da / Num. of mol.: 2 / Source method: isolated from a natural source Type: Lipoglycopeptide / Class: Antibiotic / Mass: 660.673 Da / Num. of mol.: 2 / Source method: isolated from a natural sourceDetails: BAL4850C IS A VARIANT OF THE LIPOGLYCO ARYLOMYCIN HERE BAL4850C IS REPRESENTED BY GROUPING TOGETHER THE SEQUENCE, THE LIPID (O2U) AND RHAMNOSE. Source: (natural)  Streptomyces sp. (bacteria) / References: Arylomycin Streptomyces sp. (bacteria) / References: Arylomycin#3: Chemical | 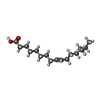  Type: Lipoglycopeptide / Class: Antibiotic / Mass: 268.435 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C17H32O2 Type: Lipoglycopeptide / Class: Antibiotic / Mass: 268.435 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C17H32O2Details: BAL4850C IS A VARIANT OF THE LIPOGLYCO ARYLOMYCIN HERE BAL4850C IS REPRESENTED BY GROUPING TOGETHER THE SEQUENCE, THE LIPID (O2U) AND RHAMNOSE. References: Arylomycin #4: Sugar |   Type: L-saccharide, alpha linking, Lipoglycopeptide / Class: Antibiotic / Mass: 164.156 Da / Num. of mol.: 2 Type: L-saccharide, alpha linking, Lipoglycopeptide / Class: Antibiotic / Mass: 164.156 Da / Num. of mol.: 2Source method: isolated from a genetically manipulated source Formula: C6H12O5 Details: BAL4850C IS A VARIANT OF THE LIPOGLYCO ARYLOMYCIN HERE BAL4850C IS REPRESENTED BY GROUPING TOGETHER THE SEQUENCE, THE LIPID (O2U) AND RHAMNOSE. References: Arylomycin #5: Water | ChemComp-HOH / | Compound details | THE GLYCO-ARYLOMYCIN IS A CYCLIC LIPOHEXAPEPTIDE, A MEMBER OF THE ARYLOMYCIN FAMILY. ALL MEMBERS ...THE GLYCO-ARYLOMYCIN | Sequence details | RESIDUE 02V of CHAIN I AND J IS N-METHYL-DIHYDROXYP | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.94 Å3/Da / Density % sol: 58.1 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7.4 Details: 22% PEG 4000, 0.2M KCl, 0.025M n-dodecyl beta-D-maltoside (DDM), pH 7.4, VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE / Date: Aug 19, 2005 / Details: mirrors |
| Radiation | Monochromator: Osimic Confocal VariMax High Flux optics / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.44→32.2 Å / Num. all: 26783 / Num. obs: 26680 / % possible obs: 99.6 % / Observed criterion σ(I): 5 / Redundancy: 9.29 % / Rmerge(I) obs: 0.105 / Net I/σ(I): 10.4 |
| Reflection shell | Resolution: 2.44→2.53 Å / Redundancy: 10.89 % / Rmerge(I) obs: 0.437 / Mean I/σ(I) obs: 5.1 / Num. unique all: 2605 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1T7D Resolution: 2.44→32.2 Å / Cor.coef. Fo:Fc: 0.915 / Cor.coef. Fo:Fc free: 0.921 / SU B: 18.315 / SU ML: 0.195 / Cross valid method: THROUGHOUT / ESU R Free: 0.253 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 56.028 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.44→32.2 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.44→2.503 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj
