+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FZD3 in inactive state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | FZD3 / complex. / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdopaminergic neuron axon guidance / serotonergic neuron axon guidance / cell proliferation in midbrain / establishment of planar polarity / negative regulation of mitotic cell cycle, embryonic / Wnt receptor activity / midbrain morphogenesis / motor neuron migration / non-canonical Wnt signaling pathway / sympathetic ganglion development ...dopaminergic neuron axon guidance / serotonergic neuron axon guidance / cell proliferation in midbrain / establishment of planar polarity / negative regulation of mitotic cell cycle, embryonic / Wnt receptor activity / midbrain morphogenesis / motor neuron migration / non-canonical Wnt signaling pathway / sympathetic ganglion development / filopodium tip / Wnt-protein binding / post-anal tail morphogenesis / negative regulation of execution phase of apoptosis / commissural neuron axon guidance / PCP/CE pathway / Class B/2 (Secretin family receptors) / Wnt signaling pathway, planar cell polarity pathway / inner ear morphogenesis / positive regulation of neuroblast proliferation / presynaptic active zone / hair follicle development / lateral plasma membrane / canonical Wnt signaling pathway / response to electrical stimulus / neural tube closure / Asymmetric localization of PCP proteins / PDZ domain binding / G protein-coupled receptor activity / electron transport chain / neuron differentiation / Ca2+ pathway / periplasmic space / electron transfer activity / iron ion binding / response to xenobiotic stimulus / apical plasma membrane / axon / neuronal cell body / dendrite / heme binding / cell surface / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
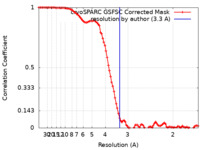
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Xu F / Zhang Z | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways. Authors: Zhibin Zhang / Xi Lin / Ling Wei / Yiran Wu / Lu Xu / Lijie Wu / Xiaohu Wei / Suwen Zhao / Xiangjia Zhu / Fei Xu /  Abstract: The ten Frizzled receptors (FZDs) are essential in Wnt signaling and play important roles in embryonic development and tumorigenesis. Among these, FZD6 is closely associated with lens development. ...The ten Frizzled receptors (FZDs) are essential in Wnt signaling and play important roles in embryonic development and tumorigenesis. Among these, FZD6 is closely associated with lens development. Understanding FZD activation mechanism is key to unlock these emerging targets. Here we present the cryo-EM structures of FZD6 and FZD3 which are known to relay non-canonical planar cell polarity (PCP) signaling pathways as well as FZD1 in their G protein-coupled states and in the apo inactive states, respectively. Comparison of the three inactive/active pairs unveiled a shared activation framework among all ten FZDs. Mutagenesis along with imaging and functional analysis on the human lens epithelial tissues suggested potential crosstalk between the G-protein coupling of FZD6 and the PCP signaling pathways. Together, this study provides an integrated understanding of FZD structure and function, and lays the foundation for developing therapeutic modulators to activate or inhibit FZD signaling for a range of disorders including cancers and cataracts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36262.map.gz emd_36262.map.gz | 79.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36262-v30.xml emd-36262-v30.xml emd-36262.xml emd-36262.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36262_fsc.xml emd_36262_fsc.xml | 9.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_36262.png emd_36262.png | 24 KB | ||
| Filedesc metadata |  emd-36262.cif.gz emd-36262.cif.gz | 6.3 KB | ||
| Others |  emd_36262_additional_1.map.gz emd_36262_additional_1.map.gz emd_36262_additional_2.map.gz emd_36262_additional_2.map.gz emd_36262_half_map_1.map.gz emd_36262_half_map_1.map.gz emd_36262_half_map_2.map.gz emd_36262_half_map_2.map.gz | 23.5 MB 45.3 MB 84.5 MB 84.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36262 http://ftp.pdbj.org/pub/emdb/structures/EMD-36262 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36262 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36262 | HTTPS FTP |
-Validation report
| Summary document |  emd_36262_validation.pdf.gz emd_36262_validation.pdf.gz | 946.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36262_full_validation.pdf.gz emd_36262_full_validation.pdf.gz | 946.1 KB | Display | |
| Data in XML |  emd_36262_validation.xml.gz emd_36262_validation.xml.gz | 17.8 KB | Display | |
| Data in CIF |  emd_36262_validation.cif.gz emd_36262_validation.cif.gz | 22.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36262 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36262 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36262 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36262 | HTTPS FTP |
-Related structure data
| Related structure data |  8jhcMC  8j9nC  8j9oC  8jh7C  8jhbC  8jhiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36262.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36262.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: mask used in the last local refine.
| File | emd_36262_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mask used in the last local refine. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: default sharpened density map after the last local refine
| File | emd_36262_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | default sharpened density map after the last local refine | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36262_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36262_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : complex of FZD3 with anti-Bril Fab and anti-Fab Nanobody
| Entire | Name: complex of FZD3 with anti-Bril Fab and anti-Fab Nanobody |
|---|---|
| Components |
|
-Supramolecule #1: complex of FZD3 with anti-Bril Fab and anti-Fab Nanobody
| Supramolecule | Name: complex of FZD3 with anti-Bril Fab and anti-Fab Nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Frizzled-3,Soluble cytochrome b562
| Macromolecule | Name: Frizzled-3,Soluble cytochrome b562 / type: protein_or_peptide / ID: 1 / Details: inactive FZD3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 74.095141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHENLYFQ GHSLFSCEPI TLRMCQDLPY NTTFMPNLLN HYDQQTAALA MEPFHPMVN LDCSRDFRPF LCALYAPICM EYGRVTLPCR RLCQRAYSEC SKLMEMFGVP WPEDMECSRF PDCDEPYPRL V DLNLAGEP ...String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHENLYFQ GHSLFSCEPI TLRMCQDLPY NTTFMPNLLN HYDQQTAALA MEPFHPMVN LDCSRDFRPF LCALYAPICM EYGRVTLPCR RLCQRAYSEC SKLMEMFGVP WPEDMECSRF PDCDEPYPRL V DLNLAGEP TEGAPVAVQR DYGFWCPREL KIDPDLGYSF LHVRDCSPPC PNMYFRREEL SFARYFIGLI SIICLSATLF TF LTFLIDV TRFRYPERPI IFYAVCYMMV SLIFFIGFLL EDRVACNASI PAQYKASTVT QGSHNKACTM LFMILYFFTM AGS VWWVIL TITWFLAAVP KWGSEAIEKK ALLFHASAWG IPGTLTIILL AMNKIEGDNI SGVCFVGLYD VDALRYFVLA PLCL YVVVG VSLLLAGIIS LNRRIARRQL ADLEDNWETL NDNLKVIEKA DNAAQVKDAL TKMRAAALDA QKATPPKLED KSPDS PEMK DFRHGFDILV GQIDDALKLA NEGKVKEAQA AAEQLKTTRN AYIQKYLERA RSTLNQDKLV KFMIRIGVFS ILYLVP LLV VIGCYFYEQA YRGIWETTWI QERCREYHIP CPYQVTQMSR PDLILFLMKY LMALIVGIPS VFWVGSKKTC FEWASFF HG R UniProtKB: Frizzled-3, Soluble cytochrome b562, Frizzled-3 |
-Macromolecule #2: anti-BRIL Fab Heavy chain
| Macromolecule | Name: anti-BRIL Fab Heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.704648 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFNVV DFSLHWVRQA PGKGLEWVAY ISSSSGSTSY ADSVKGRFTI SADTSKNTAY LQMNSLRAE DTAVYYCARW GYWPGEPWWK AFDYWGQGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ...String: EVQLVESGGG LVQPGGSLRL SCAASGFNVV DFSLHWVRQA PGKGLEWVAY ISSSSGSTSY ADSVKGRFTI SADTSKNTAY LQMNSLRAE DTAVYYCARW GYWPGEPWWK AFDYWGQGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VNHKPSNTKV DKKVEPKSLE VLFQGPHHHH HH |
-Macromolecule #3: anti-Fab Nanobody
| Macromolecule | Name: anti-Fab Nanobody / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.927301 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHENLY FQGQVQLQES GGGLVQPGGS LRLSCAASGR TISRYAMSWF RQAPGKEREF VAVARRSGDG AFYADSVQGR FTVSRDDAK NTVYLQMNSL KPEDTAVYYC AIDSDTFYSG SYDYWGQGTQ VTVSS |
-Macromolecule #4: anti-BRIL Fab Light chain
| Macromolecule | Name: anti-BRIL Fab Light chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.20982 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGTD FTLTISSLQP EDFATYYCQ QYLYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGTD FTLTISSLQP EDFATYYCQ QYLYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DIFFRACTION / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)