+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FZD6 Gs complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | FZD6 / Complex. / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationembryonic nail plate morphogenesis / establishment of body hair planar orientation / cell proliferation in midbrain / adenylate cyclase-activating serotonin receptor signaling pathway / Signaling by RNF43 mutants / midbrain morphogenesis / Wnt receptor activity / non-canonical Wnt signaling pathway / Wnt-protein binding / sensory perception of chemical stimulus ...embryonic nail plate morphogenesis / establishment of body hair planar orientation / cell proliferation in midbrain / adenylate cyclase-activating serotonin receptor signaling pathway / Signaling by RNF43 mutants / midbrain morphogenesis / Wnt receptor activity / non-canonical Wnt signaling pathway / Wnt-protein binding / sensory perception of chemical stimulus / apicolateral plasma membrane / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / PCP/CE pathway / Class B/2 (Secretin family receptors) / beta-2 adrenergic receptor binding / Wnt signaling pathway, planar cell polarity pathway / inner ear morphogenesis / PKA activation in glucagon signalling / developmental growth / hair follicle development / canonical Wnt signaling pathway / D1 dopamine receptor binding / Hedgehog 'off' state / insulin-like growth factor receptor binding / response to prostaglandin E / ionotropic glutamate receptor binding / adenylate cyclase regulator activity / Regulation of FZD by ubiquitination / cytoplasmic vesicle membrane / adenylate cyclase activator activity / neural tube closure / negative regulation of canonical Wnt signaling pathway / bone development / G protein-coupled receptor activity / platelet activation / platelet aggregation / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / cilium / ciliary basal body / apical plasma membrane / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / ubiquitin protein ligase binding / synapse Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
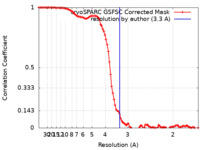
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Xu F / Zhang Z | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways. Authors: Zhibin Zhang / Xi Lin / Ling Wei / Yiran Wu / Lu Xu / Lijie Wu / Xiaohu Wei / Suwen Zhao / Xiangjia Zhu / Fei Xu /  Abstract: The ten Frizzled receptors (FZDs) are essential in Wnt signaling and play important roles in embryonic development and tumorigenesis. Among these, FZD6 is closely associated with lens development. ...The ten Frizzled receptors (FZDs) are essential in Wnt signaling and play important roles in embryonic development and tumorigenesis. Among these, FZD6 is closely associated with lens development. Understanding FZD activation mechanism is key to unlock these emerging targets. Here we present the cryo-EM structures of FZD6 and FZD3 which are known to relay non-canonical planar cell polarity (PCP) signaling pathways as well as FZD1 in their G protein-coupled states and in the apo inactive states, respectively. Comparison of the three inactive/active pairs unveiled a shared activation framework among all ten FZDs. Mutagenesis along with imaging and functional analysis on the human lens epithelial tissues suggested potential crosstalk between the G-protein coupling of FZD6 and the PCP signaling pathways. Together, this study provides an integrated understanding of FZD structure and function, and lays the foundation for developing therapeutic modulators to activate or inhibit FZD signaling for a range of disorders including cancers and cataracts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36261.map.gz emd_36261.map.gz | 80.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36261-v30.xml emd-36261-v30.xml emd-36261.xml emd-36261.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36261_fsc.xml emd_36261_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_36261.png emd_36261.png | 44.7 KB | ||
| Filedesc metadata |  emd-36261.cif.gz emd-36261.cif.gz | 6.3 KB | ||
| Others |  emd_36261_additional_1.map.gz emd_36261_additional_1.map.gz emd_36261_half_map_1.map.gz emd_36261_half_map_1.map.gz emd_36261_half_map_2.map.gz emd_36261_half_map_2.map.gz | 45.1 MB 84.7 MB 84.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36261 http://ftp.pdbj.org/pub/emdb/structures/EMD-36261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36261 | HTTPS FTP |
-Related structure data
| Related structure data |  8jhbMC  8j9nC  8j9oC  8jh7C  8jhcC  8jhiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36261.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36261.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_36261_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36261_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36261_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : FZD6-Gs complex with Nb35.
| Entire | Name: FZD6-Gs complex with Nb35. |
|---|---|
| Components |
|
-Supramolecule #1: FZD6-Gs complex with Nb35.
| Supramolecule | Name: FZD6-Gs complex with Nb35. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas type: protein_or_peptide / ID: 1 / Details: minGas / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.117201 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHENLY FQGSKTEDKR AQKRAEKKRS KLIDKQLQDE KMGYMCTHRL LLLGADNSGK STIVKQMRIL HGGSGGSGGT SGIFETKFQ VDKVNFHMFD VGGQRDERRK WIQCFNDVTA IIFVVDSSDY GSGGSGAGSA NRLQEALNLF KSIWNNRWLR T ISVILFLN ...String: HHHHHHENLY FQGSKTEDKR AQKRAEKKRS KLIDKQLQDE KMGYMCTHRL LLLGADNSGK STIVKQMRIL HGGSGGSGGT SGIFETKFQ VDKVNFHMFD VGGQRDERRK WIQCFNDVTA IIFVVDSSDY GSGGSGAGSA NRLQEALNLF KSIWNNRWLR T ISVILFLN KQDLLAEKVL AGKSKIEDYF PEFARYTTPE DATPEPGEDP RVTRAKYFIR DEFLRISTAS GDGRHYCYPH FT CAVDTEN ARRIFNDCRD IIQRMHLRQY ELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.417766 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHNTA SIAQARKLVE QLKMEANIDR IKVSKAAADL MAYCEAHAKE DPLLTPVPAS ENPFREKKFF CAIL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.845516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQVQLQESGG GLVQPGGSLR LSCAASGFTF SNYKMNWVRQ APGKGLEWVS DISQSGASIS YTGSVKGRFT ISRDNAKNTL YLQMNSLKP EDTAVYYCAR CPAPFTRDCF DVTSTTYAYR GQGTQVTVSS HHHHHH |
-Macromolecule #5: Frizzled-6
| Macromolecule | Name: Frizzled-6 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.912398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HSLFTCEPIT VPRCMKMAYN MTFFPNLMGH YDQSIAAVEM EHFLPLANLE CSPNIETFL CKAFVPTCIE QIHVVPPCRK LCEKVYSDCK KLIDTFGIRW PEELECDRLQ YCDETVPVTF DPHTEFLGPQ K KTEQVQRD ...String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HSLFTCEPIT VPRCMKMAYN MTFFPNLMGH YDQSIAAVEM EHFLPLANLE CSPNIETFL CKAFVPTCIE QIHVVPPCRK LCEKVYSDCK KLIDTFGIRW PEELECDRLQ YCDETVPVTF DPHTEFLGPQ K KTEQVQRD IGFWCPRHLK TSGGQGYKFL GIDQCAPPCP NMYFKSDELE FAKSFIGTVS IFCLCATLFT FLTFLIDVRR FR YPERPII YYSVCYSIVS LMYFIGFLLG DSTACNKADE KLELGDTVVL GSQNKACTVL FMLLYFFTMA GTVWWVILTI TWF LAAGRK WSCEAIEQKA VWFHAVAWGT PGFLTVMLLA MNKVEGDNIS GVCFVGLYDL DASRYFVLLP LCLCVFVGLS LLLA GIISL NHVRQVIQHD GRNQEKLKKF MIRIGVFSGL YLVPLVTLLG CYVYEQVNRI TWEITWVSDH CRQYHIPCPY QAKAK ARPE LALFMIKYLM TLIVGISAVF WVGSKKTCTE WAGFFKRNRK RDPISESRRV LQE UniProtKB: Frizzled-6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DIFFRACTION / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)