[English] 日本語
 Yorodumi
Yorodumi- EMDB-3305: Structure of human TFIID-IIA bound to core promoter DNA (locally-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3305 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human TFIID-IIA bound to core promoter DNA (locally-refined BC core) | |||||||||
 Map data Map data | Locally-refined BC-core of human TFIID-IIA bound to super core promoter DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TFIID / TFIIA / transcription / RNA polymerase II / general transcription factors / preinitiation complex / core promoter / DNA binding | |||||||||
| Function / homology |  Function and homology information Function and homology informationspermine transport / negative regulation of MHC class I biosynthetic process / DNA-templated transcription open complex formation / TFIIH-class transcription factor complex binding / negative regulation of protein autoubiquitination / transcription factor TFTC complex / negative regulation of MHC class II biosynthetic process / RNA polymerase transcription factor SL1 complex / regulation of cell cycle G1/S phase transition / RNA polymerase I general transcription initiation factor activity ...spermine transport / negative regulation of MHC class I biosynthetic process / DNA-templated transcription open complex formation / TFIIH-class transcription factor complex binding / negative regulation of protein autoubiquitination / transcription factor TFTC complex / negative regulation of MHC class II biosynthetic process / RNA polymerase transcription factor SL1 complex / regulation of cell cycle G1/S phase transition / RNA polymerase I general transcription initiation factor activity / SLIK (SAGA-like) complex / RNA polymerase III general transcription initiation factor activity / RNA polymerase I core promoter sequence-specific DNA binding / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / RNA Polymerase III Abortive And Retractive Initiation / positive regulation of androgen receptor activity / maintenance of protein location in nucleus / transcription factor TFIIA complex / female germ cell nucleus / male pronucleus / female pronucleus / transcription regulator inhibitor activity / RNA polymerase II general transcription initiation factor binding / nuclear vitamin D receptor binding / regulation of fat cell differentiation / RNA Polymerase I Transcription Termination / transcription preinitiation complex / SAGA complex / nuclear thyroid hormone receptor binding / inner cell mass cell proliferation / midbrain development / cellular response to ATP / histone acetyltransferase binding / transcription factor TFIID complex / RNA polymerase II general transcription initiation factor activity / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA Polymerase I Transcription Initiation / ubiquitin conjugating enzyme activity / aryl hydrocarbon receptor binding / transcription initiation at RNA polymerase I promoter / MLL1 complex / TFIIB-class transcription factor binding / transcription by RNA polymerase III / RNA polymerase II transcribes snRNA genes / negative regulation of cell cycle / P-TEFb complex binding / positive regulation of transcription initiation by RNA polymerase II / RNA polymerase II core promoter sequence-specific DNA binding / negative regulation of ubiquitin-dependent protein catabolic process / positive regulation of intrinsic apoptotic signaling pathway / regulation of DNA repair / core promoter sequence-specific DNA binding / histone acetyltransferase activity / RNA polymerase II preinitiation complex assembly / histone acetyltransferase / RNA Polymerase II Pre-transcription Events / estrogen receptor signaling pathway / TBP-class protein binding / SIRT1 negatively regulates rRNA expression / regulation of signal transduction by p53 class mediator / male germ cell nucleus / DNA-templated transcription initiation / nuclear receptor binding / peptidyl-threonine phosphorylation / transcription initiation at RNA polymerase II promoter / RNA Polymerase I Promoter Escape / lysine-acetylated histone binding / negative regulation of protein kinase activity / mRNA transcription by RNA polymerase II / NoRC negatively regulates rRNA expression / euchromatin / B-WICH complex positively regulates rRNA expression / response to organic cyclic compound / protein polyubiquitination / cellular response to UV / G2/M transition of mitotic cell cycle / p53 binding / positive regulation of protein binding / cell junction / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / kinase activity / peptidyl-serine phosphorylation / ubiquitin-dependent protein catabolic process / spermatogenesis / DNA-binding transcription factor binding / RNA polymerase II-specific DNA-binding transcription factor binding / Estrogen-dependent gene expression / Regulation of TP53 Activity through Phosphorylation / transcription regulator complex / sequence-specific DNA binding / protein autophosphorylation / transcription by RNA polymerase II Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
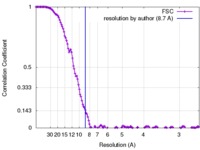
| Method | single particle reconstruction / cryo EM / Resolution: 8.7 Å | |||||||||
 Authors Authors | Louder RK / He Y / Lopez-Blanco JR / Fang J / Chacon P / Nogales E | |||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Authors: Robert K Louder / Yuan He / José Ramón López-Blanco / Jie Fang / Pablo Chacón / Eva Nogales /   Abstract: The general transcription factor IID (TFIID) plays a central role in the initiation of RNA polymerase II (Pol II)-dependent transcription by nucleating pre-initiation complex (PIC) assembly at the ...The general transcription factor IID (TFIID) plays a central role in the initiation of RNA polymerase II (Pol II)-dependent transcription by nucleating pre-initiation complex (PIC) assembly at the core promoter. TFIID comprises the TATA-binding protein (TBP) and 13 TBP-associated factors (TAF1-13), which specifically interact with a variety of core promoter DNA sequences. Here we present the structure of human TFIID in complex with TFIIA and core promoter DNA, determined by single-particle cryo-electron microscopy at sub-nanometre resolution. All core promoter elements are contacted by subunits of TFIID, with TAF1 and TAF2 mediating major interactions with the downstream promoter. TFIIA bridges the TBP-TATA complex with lobe B of TFIID. We also present the cryo-electron microscopy reconstruction of a fully assembled human TAF-less PIC. Superposition of common elements between the two structures provides novel insights into the general role of TFIID in promoter recognition, PIC assembly, and transcription initiation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3305.map.gz emd_3305.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3305-v30.xml emd-3305-v30.xml emd-3305.xml emd-3305.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3305_fsc.xml emd_3305_fsc.xml | 16 KB | Display |  FSC data file FSC data file |
| Images |  emd_3305.png emd_3305.png | 63.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3305 http://ftp.pdbj.org/pub/emdb/structures/EMD-3305 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3305 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3305 | HTTPS FTP |
-Validation report
| Summary document |  emd_3305_validation.pdf.gz emd_3305_validation.pdf.gz | 223.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3305_full_validation.pdf.gz emd_3305_full_validation.pdf.gz | 222.6 KB | Display | |
| Data in XML |  emd_3305_validation.xml.gz emd_3305_validation.xml.gz | 13.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3305 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3305 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3305 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3305 | HTTPS FTP |
-Related structure data
| Related structure data |  5furMC  3304C  3306C  3307C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3305.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3305.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally-refined BC-core of human TFIID-IIA bound to super core promoter DNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human TFIID-TFIIA complex bound to super core promoter DNA
| Entire | Name: Human TFIID-TFIIA complex bound to super core promoter DNA |
|---|---|
| Components |
|
-Supramolecule #1000: Human TFIID-TFIIA complex bound to super core promoter DNA
| Supramolecule | Name: Human TFIID-TFIIA complex bound to super core promoter DNA type: sample / ID: 1000 / Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 1.34 MDa |
-Macromolecule #1: General transcription factor IID
| Macromolecule | Name: General transcription factor IID / type: protein_or_peptide / ID: 1 / Name.synonym: TFIID / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HeLa / synonym: Human / Organelle: Nucleus / Location in cell: Nuclear extract Homo sapiens (human) / Strain: HeLa / synonym: Human / Organelle: Nucleus / Location in cell: Nuclear extract |
| Molecular weight | Theoretical: 1.26 MDa |
-Macromolecule #2: General transcription factor IIA
| Macromolecule | Name: General transcription factor IIA / type: protein_or_peptide / ID: 2 / Name.synonym: TFIIA / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 27 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #3: Super core promoter
| Macromolecule | Name: Super core promoter / type: dna / ID: 3 / Name.synonym: SCP Details: The super core promoter is a composite sequence combining promoter motifs from several strong promoters from humans and D. melanogaster. Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56 KDa |
| Sequence | String: GAAGGGCGCC TATAAAAGGG GGTGGGGGCG CGTTCGTCCT CAGTCGCGAT CGAACACTCG AGCCGAGCAG ACGTGCCTAC GGACCATGG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.9 Details: 10 mM HEPES, 10 mM MgCl2, 50 mM KCl, 3% trehalose 1 mM DTT, 0.0125% NP-40 |
| Grid | Details: Amorphous continuous carbon over C-flat holey carbon support (4 um holes with 2 um spacing) on 400 mesh copper grid. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV Method: Incubate 4 ul of sample on grid for 10 minutes, then blot for 4 seconds with force 15. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Details | The camera was operated in counting mode with a dose rate of 8 electrons/pixel per second, with a total exposure time of 10 seconds fractionated over 20 frames. |
| Date | Aug 11, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 1253 / Average electron dose: 46 e/Å2 Details: Whole-micrograph drift correction was performed using MotionCorr before averaging the frames. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 37879 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-5fur: |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-5fur: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)