+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32562 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Omicron spike protein SD1 in complex with S3H3 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / coronavirus / Omicron variant / B.1.1.529 lineage / spike protein / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / membrane fusion / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.61 Å | |||||||||
 Authors Authors | Hong Q / Cong Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Molecular basis of receptor binding and antibody neutralization of Omicron. Authors: Qin Hong / Wenyu Han / Jiawei Li / Shiqi Xu / Yifan Wang / Cong Xu / Zuyang Li / Yanxing Wang / Chao Zhang / Zhong Huang / Yao Cong /  Abstract: The SARS-CoV-2 Omicron variant exhibits striking immune evasion and is spreading rapidly worldwide. Understanding the structural basis of the high transmissibility and enhanced immune evasion of ...The SARS-CoV-2 Omicron variant exhibits striking immune evasion and is spreading rapidly worldwide. Understanding the structural basis of the high transmissibility and enhanced immune evasion of Omicron is of high importance. Here, using cryo-electron microscopy, we present both the closed and the open states of the Omicron spike (S) protein, which appear more compact than the counterparts of the G614 strain, potentially related to enhanced inter-protomer and S1-S2 interactions induced by Omicron residue substitution. The closed state showing dominant population may indicate a conformational masking mechanism for the immune evasion of Omicron. Moreover, we captured three states for the Omicron S-ACE2 complex, revealing that the substitutions on the Omicron RBM result in new salt bridges and hydrogen bonds, more favourable electrostatic surface properties, and an overall strengthened S-ACE2 interaction, in line with the observed higher ACE2 affinity of Omicron S than of G614. Furthermore, we determined the structures of Omicron S in complex with the Fab of S3H3, an antibody that is able to cross-neutralize major variants of concern including Omicron, elucidating the structural basis for S3H3-mediated broad-spectrum neutralization. Our findings shed light on the receptor engagement and antibody neutralization or evasion of Omicron and may also inform the design of broadly effective vaccines against SARS-CoV-2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32562.map.gz emd_32562.map.gz | 158.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32562-v30.xml emd-32562-v30.xml emd-32562.xml emd-32562.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32562.png emd_32562.png | 91.1 KB | ||
| Masks |  emd_32562_msk_1.map emd_32562_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32562.cif.gz emd-32562.cif.gz | 6.3 KB | ||
| Others |  emd_32562_additional_1.map.gz emd_32562_additional_1.map.gz | 89.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32562 http://ftp.pdbj.org/pub/emdb/structures/EMD-32562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32562 | HTTPS FTP |
-Validation report
| Summary document |  emd_32562_validation.pdf.gz emd_32562_validation.pdf.gz | 329.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32562_full_validation.pdf.gz emd_32562_full_validation.pdf.gz | 329 KB | Display | |
| Data in XML |  emd_32562_validation.xml.gz emd_32562_validation.xml.gz | 6.7 KB | Display | |
| Data in CIF |  emd_32562_validation.cif.gz emd_32562_validation.cif.gz | 7.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32562 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32562 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32562 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32562 | HTTPS FTP |
-Related structure data
| Related structure data |  7wk8MC  7wk2C  7wk3C  7wk4C  7wk5C  7wk6C  7wk9C  7wkaC  7wvnC  7wvoC  7wvpC  7wvqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32562.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32562.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.093 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_32562_msk_1.map emd_32562_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: SD1 S3H3 P15 J253 002 volume map
| File | emd_32562_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SD1_S3H3_P15_J253_002_volume_map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Omicron spike in complex with S3H3 Fab
| Entire | Name: SARS-CoV-2 Omicron spike in complex with S3H3 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Omicron spike in complex with S3H3 Fab
| Supramolecule | Name: SARS-CoV-2 Omicron spike in complex with S3H3 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Heavy chain of S3H3 Fab
| Macromolecule | Name: Heavy chain of S3H3 Fab / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.619521 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQQPGAE LVRPGASVKL SCKASGYSFT RFWMNWVKQR PGQGLEWIGM IHPSDSETRL NQKFKDKATL TVDKSSTTAY MQLSSPTSE DSAVYYCARK DYDYDAWFAY WGQGTLVTVS AAKTTPPSVY PLAPGSAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS ...String: QVQLQQPGAE LVRPGASVKL SCKASGYSFT RFWMNWVKQR PGQGLEWIGM IHPSDSETRL NQKFKDKATL TVDKSSTTAY MQLSSPTSE DSAVYYCARK DYDYDAWFAY WGQGTLVTVS AAKTTPPSVY PLAPGSAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS GVHTFPAVLQ SDLYTLSSSV TVPSSTWPSE TVTCNVAHPA SSTKVDKKI |
-Macromolecule #2: Light chain of S3H3 Fab
| Macromolecule | Name: Light chain of S3H3 Fab / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.5401 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVLTQSPAS LAVSLGQRAT ISCRASKSVS ASVYSYMHWY QQKPGQPPKL LIYLASSLES GVPARFSGSG SGTDFTLNIH PVEEEDAAT YYCHHSRELP PAFGGGTKLE IKRADAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT ...String: DIVLTQSPAS LAVSLGQRAT ISCRASKSVS ASVYSYMHWY QQKPGQPPKL LIYLASSLES GVPARFSGSG SGTDFTLNIH PVEEEDAAT YYCHHSRELP PAFGGGTKLE IKRADAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT DQDSKDSTYS MSSTLTLTKD EYERHNSYTC EATHKTSTSP IVKSFNR |
-Macromolecule #3: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.214625 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ PFLMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPIIVR EPEDLPQGFS ALEPLVDLPI GINITRFQTL LA LHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNITNLCPFD EVFNATRFAS VYAWNRKRIS NCVADYSVLY NLAPFFTFKC YGVSPTKLND LCFTNVYADS FVIR GDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NKLDSKVSGN YNYLYRLFRK SNLKPFERDI STEIYQAGNK PCNGV AGFN CYFPLRSYSF RPTYGVGHQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLK GTGVLTESNK KFLPFQ QFG RDIADTTDAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQGVN CTEVPVAIHA DQLTPTWRVY STGSNVF QT RAGCLIGAEY VNNSYECDIP IGAGICASYQ TQTKSHGSAS SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLK RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KYFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFKGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGA ALQIPFAMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTA SALGKLQDVV NHNAQALNTL V KQLSSKFG AISSVLNDIF SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ GSGYIPEAPR DGQAYVRKDG EWVLLSTFLE NLYFQGDYKD DDDKHHHHHH HHH UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.61 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 101192 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



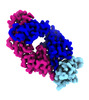

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)