[English] 日本語
 Yorodumi
Yorodumi- EMDB-31423: Cryo-EM structure of the chemokine receptor CCR5 in complex with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31423 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the chemokine receptor CCR5 in complex with RANTES and Gi | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein-coupled receptor / Chemokine receptor CCR5 / RANTES / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of chronic inflammatory response / CCR4 chemokine receptor binding / chemokine (C-C motif) ligand 5 signaling pathway / chemokine receptor antagonist activity / chemokine (C-C motif) ligand 5 binding / phospholipase D-activating G protein-coupled receptor signaling pathway / CCR1 chemokine receptor binding / positive regulation of natural killer cell chemotaxis / negative regulation of macrophage apoptotic process / chemokine receptor binding ...regulation of chronic inflammatory response / CCR4 chemokine receptor binding / chemokine (C-C motif) ligand 5 signaling pathway / chemokine receptor antagonist activity / chemokine (C-C motif) ligand 5 binding / phospholipase D-activating G protein-coupled receptor signaling pathway / CCR1 chemokine receptor binding / positive regulation of natural killer cell chemotaxis / negative regulation of macrophage apoptotic process / chemokine receptor binding / positive regulation of T cell chemotaxis / receptor signaling protein tyrosine kinase activator activity / chemokine receptor activity / CCR5 chemokine receptor binding / positive regulation of receptor signaling pathway via STAT / CCR chemokine receptor binding / positive regulation of cell-cell adhesion mediated by integrin / signaling / positive regulation of homotypic cell-cell adhesion / neutrophil activation / positive regulation of G protein-coupled receptor signaling pathway / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / negative regulation of T cell apoptotic process / C-C chemokine receptor activity / positive regulation of T cell apoptotic process / chemokine-mediated signaling pathway / eosinophil chemotaxis / C-C chemokine binding / response to cholesterol / positive regulation of calcium ion transport / positive regulation of monocyte chemotaxis / cell surface receptor signaling pathway via STAT / positive regulation of innate immune response / regulation of T cell activation / chemokine activity / Chemokine receptors bind chemokines / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / positive regulation of smooth muscle cell migration / dendritic cell chemotaxis / negative regulation of G protein-coupled receptor signaling pathway / phospholipase activator activity / leukocyte cell-cell adhesion / negative regulation of viral genome replication / chemoattractant activity / positive regulation of macrophage chemotaxis / Interleukin-10 signaling / macrophage chemotaxis / exocytosis / monocyte chemotaxis / positive regulation of translational initiation / host-mediated suppression of viral transcription / cellular response to interleukin-1 / Binding and entry of HIV virion / positive regulation of TOR signaling / cellular defense response / positive regulation of T cell migration / positive regulation of viral genome replication / adenylate cyclase inhibitor activity / coreceptor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / cellular response to fibroblast growth factor stimulus / D2 dopamine receptor binding / response to prostaglandin E / positive regulation of T cell proliferation / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / positive regulation of smooth muscle cell proliferation / adenylate cyclase-inhibiting serotonin receptor signaling pathway / regulation of insulin secretion / positive regulation of cell adhesion / cellular response to forskolin / regulation of mitotic spindle organization / epithelial cell proliferation / positive regulation of epithelial cell proliferation / cell chemotaxis / Regulation of insulin secretion / calcium-mediated signaling / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / cellular response to virus / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / cellular response to type II interferon / response to toxic substance / response to virus / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / intracellular calcium ion homeostasis / chemotaxis / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Zhang H / Chen K | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5. Authors: Hui Zhang / Kun Chen / Qiuxiang Tan / Qiang Shao / Shuo Han / Chenhui Zhang / Cuiying Yi / Xiaojing Chu / Ya Zhu / Yechun Xu / Qiang Zhao / Beili Wu /  Abstract: The chemokine receptor CCR5 plays a vital role in immune surveillance and inflammation. However, molecular details that govern its endogenous chemokine recognition and receptor activation remain ...The chemokine receptor CCR5 plays a vital role in immune surveillance and inflammation. However, molecular details that govern its endogenous chemokine recognition and receptor activation remain elusive. Here we report three cryo-electron microscopy structures of G protein-coupled CCR5 in a ligand-free state and in complex with the chemokine MIP-1α or RANTES, as well as the crystal structure of MIP-1α-bound CCR5. These structures reveal distinct binding modes of the two chemokines and a specific accommodate pattern of the chemokine for the distal N terminus of CCR5. Together with functional data, the structures demonstrate that chemokine-induced rearrangement of toggle switch and plasticity of the receptor extracellular region are critical for receptor activation, while a conserved tryptophan residue in helix II acts as a trigger of receptor constitutive activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31423.map.gz emd_31423.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31423-v30.xml emd-31423-v30.xml emd-31423.xml emd-31423.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31423.png emd_31423.png | 32.1 KB | ||
| Filedesc metadata |  emd-31423.cif.gz emd-31423.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31423 http://ftp.pdbj.org/pub/emdb/structures/EMD-31423 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31423 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31423 | HTTPS FTP |
-Validation report
| Summary document |  emd_31423_validation.pdf.gz emd_31423_validation.pdf.gz | 513.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31423_full_validation.pdf.gz emd_31423_full_validation.pdf.gz | 513.2 KB | Display | |
| Data in XML |  emd_31423_validation.xml.gz emd_31423_validation.xml.gz | 6 KB | Display | |
| Data in CIF |  emd_31423_validation.cif.gz emd_31423_validation.cif.gz | 6.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31423 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31423 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31423 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31423 | HTTPS FTP |
-Related structure data
| Related structure data |  7f1rMC  7f1qC  7f1sC  7f1tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31423.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31423.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
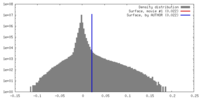
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Chemokine receptor CCR5 in complex with RANTES and Gi
| Entire | Name: Chemokine receptor CCR5 in complex with RANTES and Gi |
|---|---|
| Components |
|
-Supramolecule #1: Chemokine receptor CCR5 in complex with RANTES and Gi
| Supramolecule | Name: Chemokine receptor CCR5 in complex with RANTES and Gi / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: C-C motif chemokine 5,C-C chemokine receptor type 5
| Macromolecule | Name: C-C motif chemokine 5,C-C chemokine receptor type 5 / type: protein_or_peptide / ID: 1 / Details: Fusion protein of RANTES and CCR5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.101227 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SPYSSDTTPC CFAYIARPLP RAHIKEYCYT SGKCSNPAVV FVTRKNRQVC ANPEKKWVRE YINSLEMSEF LEGSGSGSGS GSGSGSGSG SGSGSGSDYQ VSSPIYDINY YTSEPCQKIN VKQIAARLLP PLYSLVFIFG FVGNMLVILI LINCKRLKSM T DIYLLNLA ...String: SPYSSDTTPC CFAYIARPLP RAHIKEYCYT SGKCSNPAVV FVTRKNRQVC ANPEKKWVRE YINSLEMSEF LEGSGSGSGS GSGSGSGSG SGSGSGSDYQ VSSPIYDINY YTSEPCQKIN VKQIAARLLP PLYSLVFIFG FVGNMLVILI LINCKRLKSM T DIYLLNLA ISDLFFLLTV PFWAHYAAAQ WDFGNTMCQL LTGLYFIGFF SGIFFIILLT IDRYLAVVHA VFALKARTVT FG VVTSVIT WVVAVFASLP NIIFTRSQKC GLHYTCSSHF PYSQYQFWKN FQTLKIVILG LVLPLLVMVI CYSGILKTLL RCR NEKKRH RAVRLIFTIM IVYFLFWAPY NIVLLLNTFQ EFFGLNNCSS SNRLDQAMQV TETLGMTHCC INPIIYAFVG EKFR NYLLV FFQKHIAKRE FLEVLFQGPG SWSHPQFEKG SGAGASAGSW SHPQFEKGSD YKDDDDK UniProtKB: C-C motif chemokine 5, C-C chemokine receptor type 5 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.447141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKCTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKCTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVTAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 2.1875 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1304062 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)