+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30312 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human gamma-secretase in complex with small molecule L-685,458 | |||||||||
 Map data Map data | gamma-secretase in complex with a small molecule | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / inhibitor / MEMBRANE PROTEIN / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationCajal-Retzius cell differentiation / positive regulation of L-glutamate import across plasma membrane / amyloid precursor protein biosynthetic process / negative regulation of core promoter binding / Notch receptor processing / positive regulation of endopeptidase activity / gamma-secretase complex / short-term synaptic potentiation / aspartic endopeptidase activity, intramembrane cleaving / positive regulation of amyloid precursor protein biosynthetic process ...Cajal-Retzius cell differentiation / positive regulation of L-glutamate import across plasma membrane / amyloid precursor protein biosynthetic process / negative regulation of core promoter binding / Notch receptor processing / positive regulation of endopeptidase activity / gamma-secretase complex / short-term synaptic potentiation / aspartic endopeptidase activity, intramembrane cleaving / positive regulation of amyloid precursor protein biosynthetic process / smooth endoplasmic reticulum calcium ion homeostasis / Noncanonical activation of NOTCH3 / protein catabolic process at postsynapse / TGFBR3 PTM regulation / sequestering of calcium ion / synaptic vesicle targeting / positive regulation of coagulation / central nervous system myelination / negative regulation of axonogenesis / membrane protein intracellular domain proteolysis / skin morphogenesis / choline transport / T cell activation involved in immune response / NOTCH4 Activation and Transmission of Signal to the Nucleus / dorsal/ventral neural tube patterning / ciliary rootlet / Regulated proteolysis of p75NTR / neural retina development / regulation of resting membrane potential / L-glutamate import across plasma membrane / myeloid dendritic cell differentiation / regulation of phosphorylation / metanephros development / endoplasmic reticulum calcium ion homeostasis / brain morphogenesis / amyloid precursor protein metabolic process / regulation of synaptic vesicle cycle / locomotion / regulation of long-term synaptic potentiation / regulation of postsynapse organization / embryonic limb morphogenesis / cell fate specification / astrocyte activation involved in immune response / regulation of canonical Wnt signaling pathway / myeloid cell homeostasis / aggresome / growth factor receptor binding / skeletal system morphogenesis / Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases / Golgi cisterna membrane / azurophil granule membrane / positive regulation of amyloid fibril formation / glutamate receptor signaling pathway / G protein-coupled dopamine receptor signaling pathway / amyloid precursor protein catabolic process / : / blood vessel development / amyloid-beta formation / mitochondrial transport / heart looping / regulation of neuron projection development / positive regulation of dendritic spine development / adult behavior / cerebral cortex cell migration / membrane protein ectodomain proteolysis / smooth endoplasmic reticulum / positive regulation of receptor recycling / nuclear outer membrane / negative regulation of apoptotic signaling pathway / EPH-ephrin mediated repulsion of cells / autophagosome assembly / negative regulation of ubiquitin-dependent protein catabolic process / endopeptidase activator activity / neuron development / somitogenesis / Nuclear signaling by ERBB4 / T cell proliferation / hematopoietic progenitor cell differentiation / regulation of synaptic transmission, glutamatergic / calcium ion homeostasis / Notch signaling pathway / Degradation of the extracellular matrix / rough endoplasmic reticulum / neuron projection maintenance / astrocyte activation / NOTCH2 Activation and Transmission of Signal to the Nucleus / cellular response to calcium ion / NRIF signals cell death from the nucleus / Activated NOTCH1 Transmits Signal to the Nucleus / thymus development / cerebellum development / positive regulation of glycolytic process / epithelial cell proliferation / dendritic shaft / post-embryonic development / PDZ domain binding / neuromuscular junction / apoptotic signaling pathway / NOTCH3 Activation and Transmission of Signal to the Nucleus / cell-cell adhesion Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Yang G / Zhou R | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Structural basis of γ-secretase inhibition and modulation by small molecule drugs. Authors: Guanghui Yang / Rui Zhou / Xuefei Guo / Chuangye Yan / Jianlin Lei / Yigong Shi /  Abstract: Development of γ-secretase inhibitors (GSIs) and modulators (GSMs) represents an attractive therapeutic opportunity for Alzheimer's disease (AD) and cancers. However, how these GSIs and GSMs target ...Development of γ-secretase inhibitors (GSIs) and modulators (GSMs) represents an attractive therapeutic opportunity for Alzheimer's disease (AD) and cancers. However, how these GSIs and GSMs target γ-secretase has remained largely unknown. Here, we report the cryoelectron microscopy (cryo-EM) structures of human γ-secretase bound individually to two GSI clinical candidates, Semagacestat and Avagacestat, a transition state analog GSI L685,458, and a classic GSM E2012, at overall resolutions of 2.6-3.1 Å. Remarkably, each of the GSIs occupies the same general location on presenilin 1 (PS1) that accommodates the β strand from amyloid precursor protein or Notch, interfering with substrate recruitment. L685,458 directly coordinates the two catalytic aspartate residues of PS1. E2012 binds to an allosteric site of γ-secretase on the extracellular side, potentially explaining its modulating activity. Structural analysis reveals a set of shared themes and variations for inhibitor and modulator recognition that will guide development of the next-generation substrate-selective inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30312.map.gz emd_30312.map.gz | 116.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30312-v30.xml emd-30312-v30.xml emd-30312.xml emd-30312.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30312.png emd_30312.png | 88.8 KB | ||
| Filedesc metadata |  emd-30312.cif.gz emd-30312.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30312 http://ftp.pdbj.org/pub/emdb/structures/EMD-30312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30312 | HTTPS FTP |
-Validation report
| Summary document |  emd_30312_validation.pdf.gz emd_30312_validation.pdf.gz | 522.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30312_full_validation.pdf.gz emd_30312_full_validation.pdf.gz | 521.7 KB | Display | |
| Data in XML |  emd_30312_validation.xml.gz emd_30312_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_30312_validation.cif.gz emd_30312_validation.cif.gz | 7.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30312 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30312 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30312 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30312 | HTTPS FTP |
-Related structure data
| Related structure data |  7c9iMC  0944C  0957C  6lqgC  6lr4C  7d8xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30312.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30312.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | gamma-secretase in complex with a small molecule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.091 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human gamma-secretase
| Entire | Name: human gamma-secretase |
|---|---|
| Components |
|
-Supramolecule #1: human gamma-secretase
| Supramolecule | Name: human gamma-secretase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Nicastrin
| Macromolecule | Name: Nicastrin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 78.48357 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATAGGGSGA DPGSRGLLRL LSFCVLLAGL CRGNSVERKI YIPLNKTAPC VRLLNATHQI GCQSSISGDT GVIHVVEKEE DLQWVLTDG PNPPYMVLLE SKHFTRDLME KLKGRTSRIA GLAVSLTKPS PASGFSPSVQ CPNDGFGVYS NSYGPEFAHC R EIQWNSLG ...String: MATAGGGSGA DPGSRGLLRL LSFCVLLAGL CRGNSVERKI YIPLNKTAPC VRLLNATHQI GCQSSISGDT GVIHVVEKEE DLQWVLTDG PNPPYMVLLE SKHFTRDLME KLKGRTSRIA GLAVSLTKPS PASGFSPSVQ CPNDGFGVYS NSYGPEFAHC R EIQWNSLG NGLAYEDFSF PIFLLEDENE TKVIKQCYQD HNLSQNGSAP TFPLCAMQLF SHMHAVISTA TCMRRSSIQS TF SINPEIV CDPLSDYNVW SMLKPINTTG TLKPDDRVVV AATRLDSRSF FWNVAPGAES AVASFVTQLA AAEALQKAPD VTT LPRNVM FVFFQGETFD YIGSSRMVYD MEKGKFPVQL ENVDSFVELG QVALRTSLEL WMHTDPVSQK NESVRNQVED LLAT LEKSG AGVPAVILRR PNQSQPLPPS SLQRFLRARN ISGVVLADHS GAFHNKYYQS IYDTAENINV SYPEWLSPEE DLNFV TDTA KALADVATVL GRALYELAGG TNFSDTVQAD PQTVTRLLYG FLIKANNSWF QSILRQDLRS YLGDGPLQHY IAVSSP TNT TYVVQYALAN LTGTVVNLTR EQCQDPSKVP SENKDLYEYS WVQGPLHSNE TDRLPRCVRS TARLARALSP AFELSQW SS TEYSTWTESR WKDIRARIFL IASKELELIT LTVGFGILIF SLIVTYCINA KADVLFIAPR EPGAVSY UniProtKB: Nicastrin |
-Macromolecule #2: Presenilin-1
| Macromolecule | Name: Presenilin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.713535 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTELPAPLSY FQNAQMSEDN HLSNTVRSQN DNRERQEHND RRSLGHPEPL SNGRPQGNSR QVVEQDEEED EELTLKYGAK HVIMLFVPV TLCMVVVVAT IKSVSFYTRK DGQLIYTPFT EDTETVGQRA LHSILNAAIM ISVIVVMTIL LVVLYKYRCY K VIHAWLII ...String: MTELPAPLSY FQNAQMSEDN HLSNTVRSQN DNRERQEHND RRSLGHPEPL SNGRPQGNSR QVVEQDEEED EELTLKYGAK HVIMLFVPV TLCMVVVVAT IKSVSFYTRK DGQLIYTPFT EDTETVGQRA LHSILNAAIM ISVIVVMTIL LVVLYKYRCY K VIHAWLII SSLLLLFFFS FIYLGEVFKT YNVAVDYITV ALLIWNFGVV GMISIHWKGP LRLQQAYLIM ISALMALVFI KY LPEWTAW LILAVISVYD LVAVLCPKGP LRMLVETAQE RNETLFPALI YSSTMVWLVN MAEGDPEAQR RVSKNSKYNA EST ERESQD TVAENDDGGF SEEWEAQRDS HLGPHRSTPE SRAAVQELSS SILAGEDPEE RGVKLGLGDF IFYSVLVGKA SATA SGDWN TTIACFVAIL IGLCLTLLLL AIFKKALPAL PISITFGLVF YFATDYLVQP FMDQLAFHQF YI UniProtKB: Presenilin-1 |
-Macromolecule #3: Gamma-secretase subunit APH-1A
| Macromolecule | Name: Gamma-secretase subunit APH-1A / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.017943 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAAVFFGCT FVAFGPAFAL FLITVAGDPL RVIILVAGAF FWLVSLLLAS VVWFILVHVT DRSDARLQYG LLIFGAAVSV LLQEVFRFA YYKLLKKADE GLASLSEDGR SPISIRQMAY VSGLSFGIIS GVFSVINILA DALGPGVVGI HGDSPYYFLT S AFLTAAII ...String: MGAAVFFGCT FVAFGPAFAL FLITVAGDPL RVIILVAGAF FWLVSLLLAS VVWFILVHVT DRSDARLQYG LLIFGAAVSV LLQEVFRFA YYKLLKKADE GLASLSEDGR SPISIRQMAY VSGLSFGIIS GVFSVINILA DALGPGVVGI HGDSPYYFLT S AFLTAAII LLHTFWGVVF FDACERRRYW ALGLVVGSHL LTSGLTFLNP WYEASLLPIY AVTVSMGLWA FITAGGSLRS IQ RSLLCRR QEDSRVMVYS ALRIPPED UniProtKB: Gamma-secretase subunit APH-1A |
-Macromolecule #4: Gamma-secretase subunit PEN-2
| Macromolecule | Name: Gamma-secretase subunit PEN-2 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.49868 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASWSHPQFE KGGGARGGSG GGSWSHPQFE KGFDYKDDDD KGTNLERVSN EEKLNLCRKY YLGGFAFLPF LWLVNIFWFF REAFLVPAY TEQSQIKGYV WRSAVGFLFW VIVLTSWITI FQIYRPRWGA LGDYLSFTIP LGTP UniProtKB: Gamma-secretase subunit PEN-2 |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 8 / Number of copies: 2 / Formula: PC1 |
|---|---|
| Molecular weight | Theoretical: 790.145 Da |
| Chemical component information |  ChemComp-PC1: |
-Macromolecule #9: ~{tert}-butyl ~{N}-[(2~{S},3~{R},5~{R})-6-[[(2~{S})-1-[[(2~{S})-1...
| Macromolecule | Name: ~{tert}-butyl ~{N}-[(2~{S},3~{R},5~{R})-6-[[(2~{S})-1-[[(2~{S})-1-azanyl-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-oxidanyl-6-oxidanylidene-1- ...Name: ~{tert}-butyl ~{N}-[(2~{S},3~{R},5~{R})-6-[[(2~{S})-1-[[(2~{S})-1-azanyl-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-oxidanyl-6-oxidanylidene-1-phenyl-5-(phenylmethyl)hexan-2-yl]carbamate type: ligand / ID: 9 / Number of copies: 1 / Formula: FTO |
|---|---|
| Molecular weight | Theoretical: 672.853 Da |
| Chemical component information | 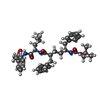 ChemComp-FTO: |
-Macromolecule #10: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 10 / Number of copies: 3 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.5625 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















