[English] 日本語
 Yorodumi
Yorodumi- EMDB-30008: Cryo-EM structure of human secretory immunoglobulin A in complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30008 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human secretory immunoglobulin A in complex with the N-terminal domain of SpsA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | immunoglobulin / dimer / transcytosis / secreted / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationpolymeric immunoglobulin receptor activity / immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor / kappa-type opioid receptor binding / response to tacrolimus / polymeric immunoglobulin binding / regulation of T cell homeostatic proliferation / dimeric IgA immunoglobulin complex / regulation of CD4-positive, alpha-beta T cell proliferation / interleukin-2 receptor binding / secretory dimeric IgA immunoglobulin complex ...polymeric immunoglobulin receptor activity / immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor / kappa-type opioid receptor binding / response to tacrolimus / polymeric immunoglobulin binding / regulation of T cell homeostatic proliferation / dimeric IgA immunoglobulin complex / regulation of CD4-positive, alpha-beta T cell proliferation / interleukin-2 receptor binding / secretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / secretory IgA immunoglobulin complex / glycosphingolipid binding / positive regulation of plasma cell differentiation / negative regulation of lymphocyte proliferation / Fc receptor signaling pathway / IgA binding / positive regulation of tissue remodeling / negative regulation of T-helper 17 cell differentiation / IgA immunoglobulin complex / RUNX1 and FOXP3 control the development of regulatory T lymphocytes (Tregs) / glomerular filtration / leukocyte activation involved in immune response / detection of chemical stimulus involved in sensory perception of bitter taste / positive regulation of isotype switching to IgG isotypes / interleukin-2-mediated signaling pathway / activated T cell proliferation / IgG immunoglobulin complex / cell surface receptor signaling pathway via STAT / Interleukin-2 signaling / natural killer cell activation / kinase activator activity / immunoglobulin receptor binding / positive regulation of regulatory T cell differentiation / immunoglobulin complex, circulating / negative regulation of B cell apoptotic process / positive regulation of immunoglobulin production / azurophil granule membrane / receptor clustering / positive regulation of dendritic spine development / positive regulation of interleukin-17 production / positive regulation of respiratory burst / humoral immune response / positive regulation of activated T cell proliferation / T cell differentiation / Interleukin receptor SHC signaling / complement activation, classical pathway / Scavenging of heme from plasma / antigen binding / positive regulation of B cell proliferation / extrinsic apoptotic signaling pathway in absence of ligand / cytokine activity / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / growth factor activity / negative regulation of inflammatory response / epidermal growth factor receptor signaling pathway / positive regulation of type II interferon production / positive regulation of inflammatory response / transmembrane signaling receptor activity / antibacterial humoral response / cell-cell signaling / positive regulation of cytosolic calcium ion concentration / carbohydrate binding / RAF/MAP kinase cascade / positive regulation of cell growth / protein-containing complex assembly / response to ethanol / blood microparticle / protein-macromolecule adaptor activity / phospholipase C-activating G protein-coupled receptor signaling pathway / adaptive immune response / transcription by RNA polymerase II / receptor complex / cell adhesion / immune response / innate immune response / positive regulation of cell population proliferation / Neutrophil degranulation / negative regulation of apoptotic process / signal transduction / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
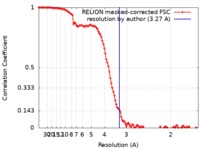
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Wang Y / Wang G | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2020 Journal: Cell Res / Year: 2020Title: Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin. Authors: Yuxin Wang / Guopeng Wang / Yaxin Li / Qinyu Zhu / Hao Shen / Ning Gao / Junyu Xiao /  Abstract: Secretory Immunoglobulin A (SIgA) is the most abundant antibody at the mucosal surface. It possesses two additional subunits besides IgA: the joining chain (J-chain) and secretory component (SC). SC ...Secretory Immunoglobulin A (SIgA) is the most abundant antibody at the mucosal surface. It possesses two additional subunits besides IgA: the joining chain (J-chain) and secretory component (SC). SC is the ectodomain of the polymeric immunoglobulin receptor (pIgR), which functions to transport IgA to the mucosa. How the J-chain and pIgR/SC facilitate the assembly and secretion of SIgA remains incompletely understood. Furthermore, during the infection of Streptococcus pneumoniae, the pneumococcal adhesin SpsA hijacks pIgR/SC and SIgA to gain entry to human cells and evade host defense. How SpsA targets pIgR/SC and SIgA also remains elusive. Here we report a cryo-electron microscopy structure of the Fc region of IgA1 (Fcα) in complex with the J-chain and SC (Fcα-J-SC), which reveals the organization principle of SIgA. We also present a structure of Fcα-J-SC complexed with SpsA, which uncovers the specific interactions between SpsA and human pIgR/SC. These results advance the molecular understanding of SIgA and shed light on S. pneumoniae pathogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30008.map.gz emd_30008.map.gz | 9.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30008-v30.xml emd-30008-v30.xml emd-30008.xml emd-30008.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30008_fsc.xml emd_30008_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_30008.png emd_30008.png | 49.7 KB | ||
| Masks |  emd_30008_msk_1.map emd_30008_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-30008.cif.gz emd-30008.cif.gz | 6.8 KB | ||
| Others |  emd_30008_half_map_1.map.gz emd_30008_half_map_1.map.gz emd_30008_half_map_2.map.gz emd_30008_half_map_2.map.gz | 98.2 MB 98.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30008 http://ftp.pdbj.org/pub/emdb/structures/EMD-30008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30008 | HTTPS FTP |
-Related structure data
| Related structure data |  6lxwMC  6lx3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30008.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30008.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_30008_msk_1.map emd_30008_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_30008_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_30008_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Quadruple complex of human secretory immunoglobulin A with the N-...
| Entire | Name: Quadruple complex of human secretory immunoglobulin A with the N-terminal domain of SpsA |
|---|---|
| Components |
|
-Supramolecule #1: Quadruple complex of human secretory immunoglobulin A with the N-...
| Supramolecule | Name: Quadruple complex of human secretory immunoglobulin A with the N-terminal domain of SpsA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Interleukin-2,Immunoglobulin heavy constant alpha 1
| Macromolecule | Name: Interleukin-2,Immunoglobulin heavy constant alpha 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.445693 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYRMQLLSCI ALSLALVTNS ARIHMSAWSH PQFEKGGGSG GGSGGSAWSH PQFEKIDTTC CHPRLSLHRP ALEDLLLGSE ANLTCTLTG LRDASGVTFT WTPSSGKSAV QGPPERDLCG CYSVSSVLPG CAEPWNHGKT FTCTAAYPES KTPLTATLSK S GNTFRPEV ...String: MYRMQLLSCI ALSLALVTNS ARIHMSAWSH PQFEKGGGSG GGSGGSAWSH PQFEKIDTTC CHPRLSLHRP ALEDLLLGSE ANLTCTLTG LRDASGVTFT WTPSSGKSAV QGPPERDLCG CYSVSSVLPG CAEPWNHGKT FTCTAAYPES KTPLTATLSK S GNTFRPEV HLLPPPSEEL ALNELVTLTC LARGFSPKDV LVRWLQGSQE LPREKYLTWA SRQEPSQGTT TFAVTSILRV AA EDWKKGD TFSCMVGHEA LPLAFTQKTI DRLAGKPTHV NVSVVMAEVD GTCY UniProtKB: Interleukin-2, Immunoglobulin heavy constant alpha 1 |
-Macromolecule #2: Immunoglobulin J chain
| Macromolecule | Name: Immunoglobulin J chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 19.225762 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKNHLLFWGV LAVFIKAVHV KAQEDERIVL VDNKCKCARI TSRIIRSSED PNEDIVERNI RIIVPLNNRE NISDPTSPLR TRFVYHLSD LCKKCDPTEV ELDNQIVTAT QSNICDEDSA TETCYTYDRN KCYTAVVPLV YGGETKMVET ALTPDACYPD H HHHHHHH UniProtKB: Immunoglobulin J chain |
-Macromolecule #3: Polymeric immunoglobulin receptor
| Macromolecule | Name: Polymeric immunoglobulin receptor / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.306336 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLLFVLTCLL AVFPAISTKS PIFGPEEVNS VEGNSVSITC YYPPTSVNRH TRKYWCRQGA RGGCITLISS EGYVSSKYAG RANLTNFPE NGTFVVNIAQ LSQDDSGRYK CGLGINSRGL SFDVSLEVSQ GPGLLNDTKV YTVDLGRTVT INCPFKTENA Q KRKSLYKQ ...String: MLLFVLTCLL AVFPAISTKS PIFGPEEVNS VEGNSVSITC YYPPTSVNRH TRKYWCRQGA RGGCITLISS EGYVSSKYAG RANLTNFPE NGTFVVNIAQ LSQDDSGRYK CGLGINSRGL SFDVSLEVSQ GPGLLNDTKV YTVDLGRTVT INCPFKTENA Q KRKSLYKQ IGLYPVLVID SSGYVNPNYT GRIRLDIQGT GQLLFSVVIN QLRLSDAGQY LCQAGDDSNS NKKNADLQVL KP EPELVYE DLRGSVTFHC ALGPEVANVA KFLCRQSSGE NCDVVVNTLG KRAPAFEGRI LLNPQDKDGS FSVVITGLRK EDA GRYLCG AHSDGQLQEG SPIQAWQLFV NEESTIPRSP TVVKGVAGGS VAVLCPYNRK ESKSIKYWCL WEGAQNGRCP LLVD SEGWV KAQYEGRLSL LEEPGNGTFT VILNQLTSRD AGFYWCLTNG DTLWRTTVEI KIIEGEPNLK VPGNVTAVLG ETLKV PCHF PCKFSSYEKY WCKWNNTGCQ ALPSQDEGPS KAFVNCDENS RLVSLTLNLV TRADEGWYWC GVKQGHFYGE TAAVYV AVE ERHHHHHHHH UniProtKB: Polymeric immunoglobulin receptor |
-Macromolecule #4: SigA binding protein
| Macromolecule | Name: SigA binding protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.311113 KDa |
| Recombinant expression | Organism:  Escherichia phage Ecwhy_1 (virus) Escherichia phage Ecwhy_1 (virus) |
| Sequence | String: MGSHHHHHHH HGSDYDIPTT ENLYFQGSEF TENEGSTQAA TFSNMANKSQ TEQGEINIER DKAKTAVSEY KEKKVSEIYT KLERDRHKD TVDLVNKLQE IKNEYLNKIV QSTSKTEIQG LITTSRSKLD EAVSKYKKAP SSSSSSGSST KPEASDTAKP N KPTELEKK ...String: MGSHHHHHHH HGSDYDIPTT ENLYFQGSEF TENEGSTQAA TFSNMANKSQ TEQGEINIER DKAKTAVSEY KEKKVSEIYT KLERDRHKD TVDLVNKLQE IKNEYLNKIV QSTSKTEIQG LITTSRSKLD EAVSKYKKAP SSSSSSGSST KPEASDTAKP N KPTELEKK VAEAEKKVEE AKKKAKDQKE EDYRNYPTIT YKTLELEIAE SDVEVKKAEL ELVKEEAKEP RNEEKVKQAK AK VESEETE ATRLEKIKTD RKKAEEEAKR KAAEEDKVKE KPAEQQAEED YARRSEEEYN RLTQQQPPKT EKPAQPSTPK UniProtKB: SigA binding protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 59.74 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)