+ Open data
Open data
- Basic information
Basic information
| Entry | 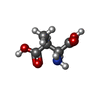 Database: PDB chemical components / ID: 2AS Database: PDB chemical components / ID: 2AS |
|---|---|
| Name | Name: ( |
-Chemical information
| Composition |  | ||||||
|---|---|---|---|---|---|---|---|
| Others | Type: L-peptide linking / PDB classification: HETAIN / One letter code: X / Three letter code: 2AS / Ideal coordinates details: Corina / Model coordinates PDB-ID: 1W3M | ||||||
| History |
| ||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.352 | OpenEye OEToolkits 1.7.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.352 | | OpenEye OEToolkits 1.7.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | (| OpenEye OEToolkits 1.6.1 | ( | |
|---|
-PDB entries
Showing all 4 items

PDB-1i9c: 
GLUTAMATE MUTASE FROM CLOSTRIDIUM COCHLEARIUM: COMPLEX WITH ADENOSYLCOBALAMIN AND SUBSTRATE

PDB-1kkr: 
CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE CONTAINING (2S,3S)-3-METHYLASPARTIC ACID

PDB-3wv5: 
Complex structure of VinN with 3-methylaspartate

PDB-5xnk: 
Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with DL-methyl-aspartate
 Movie
Movie Controller
Controller